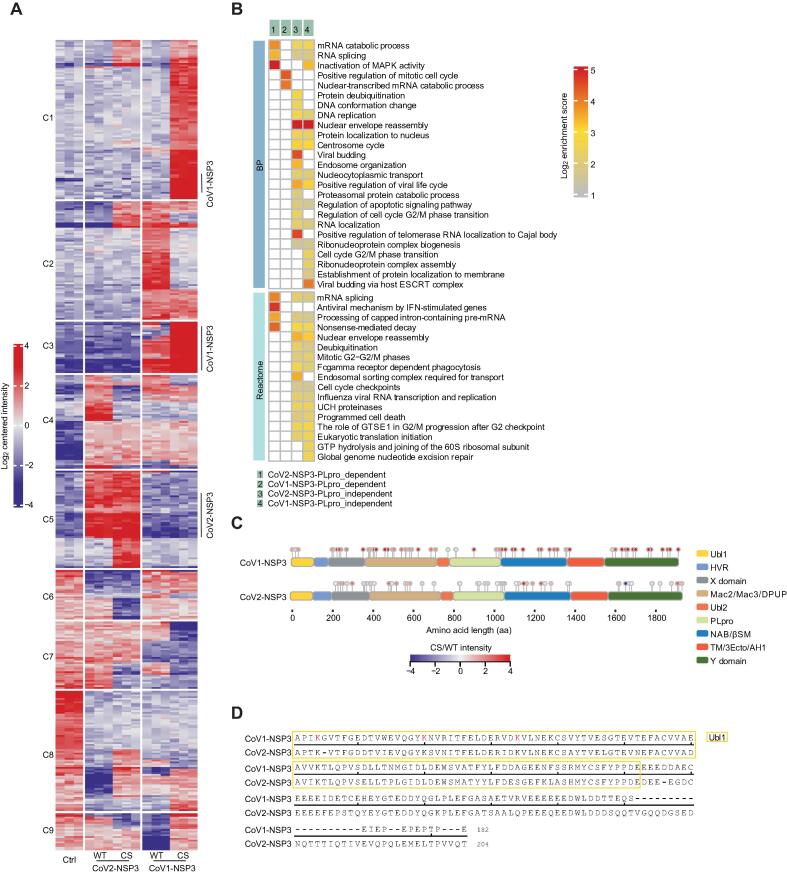
Figure 6.
Ubiquitylome analysis of NSP3-expressing cells
A. Clustering of DEUPs in NSP3-expressing cells. DEUPs showing significantly differences in NSP3-expressing cells compared to control cells were clustered together. C1–C9 on the left indicate Clusters 1–9. Full list of DEUPs is available in Table S6. B. GO BP and Reactome analyses of catalytic activity-dependent and -independent DEUPs. Full list of enriched terms is available in Table S6. C. Schematic representation of identified ubiquitination sites of NSP3 proteins. The intensity of each ubiquitinated site was log2-transformed and compared between CS-mutated and WT NSP3-expressing cells. D. Alignment of the Ubl1 and HVR domains of NSP3 proteins. Red-labeled K indicates identified ubiquitinated lysine residue of CoV1-NSP3. DEUP, differentially enriched ubiquitinated peptide.