Figure 6: Intrinsic TGF-beta signaling contributes to CAF functional heterogeneity by suppressing HGF and FGF7 expression.
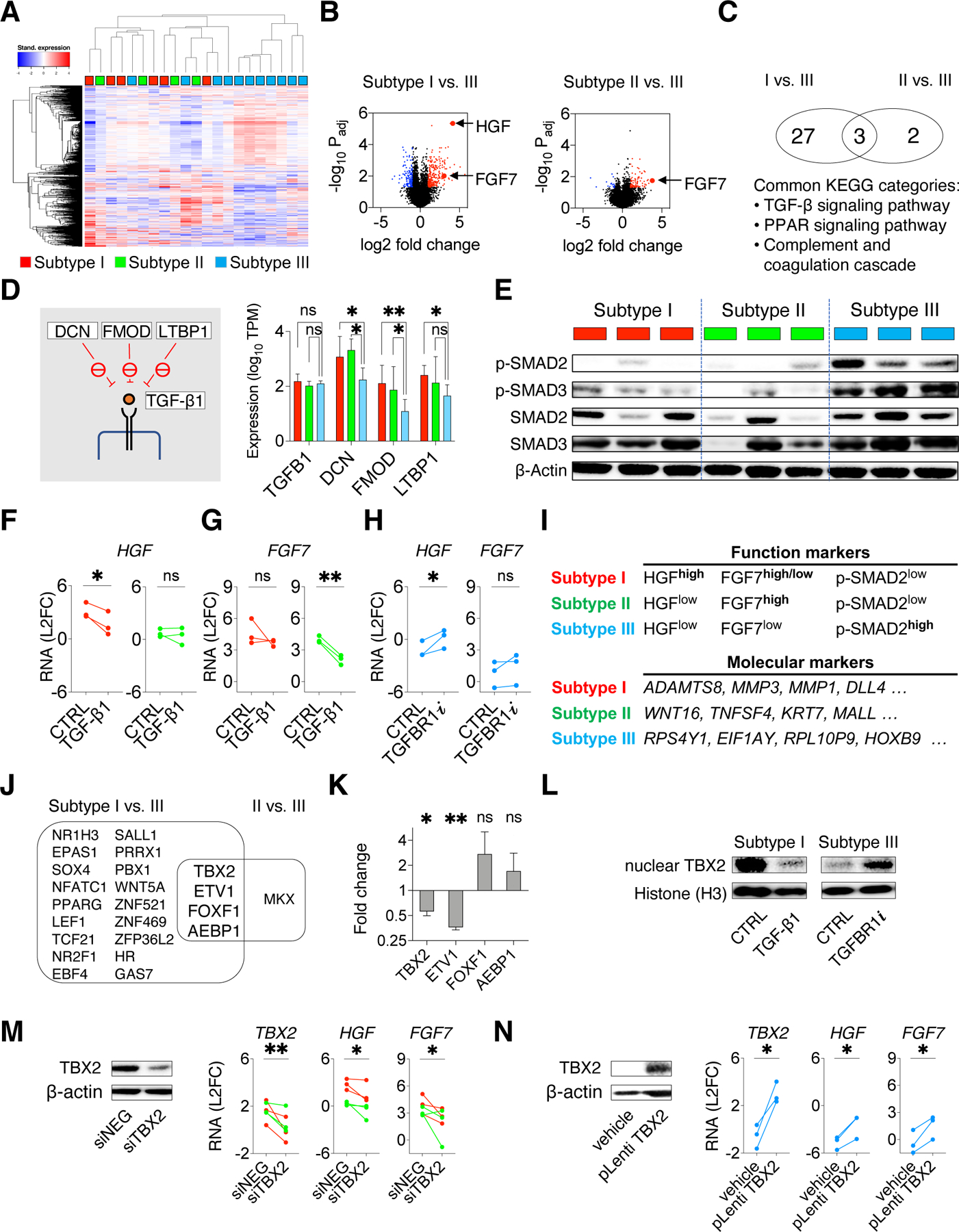
A. Heatmap with unsupervised clustering showing the top 1000 differentially expressed genes across a total of 21 PDFs. B. Volcano plots show the over-expressed genes (red) and under-expressed genes (blue) in subtype I or II PDFs compared with subtype III PDFs. C. Venn diagram showing pathways (KEGG annotation) related with genes over-expressed in subtypes I and II PDFs. D. RNAseq expressions of TGF-β1 and TGF-β1 upstream suppressors DCN, FMOD, and LTBP1(schematics on the left) in subtypes I (red), II (green), and III PDFs (blue). Mean with 95% CI. Two-tailed t test is used. E. Western blotting shows TGF-β signaling (phospho-SMAD2/SMAD3) in PDFs. Lysates were also probed in Figure S7B. F-G. HGF (F) and FGF7 (G) RNA expression measured by qRT-PCR in subtypes I (red) and II (green) PDFs upon activating TGF-β signaling using TGF-β1 (10ng/mL) for 24 hours. H. HGF and FGF7 RNA expression in subtype III (blue) PDFs after TGFBR1 inhibitor vactosertib (1μM) treatment for 24 hours. I. Function markers (HGF, FGF7, and phospho-SMAD2) and molecular markers (most variably expressed genes identified by PDF RNA sequencing, top four genes are shown) to distinguish CAF functional subtypes. J. Venn diagram shows transcription factor genes commonly over-expressed in subtype I and subtype II PDFs. K. RNA expression change of the indicated transcription factors genes in subtype I PDFs after treating with TGF-β1 for 24 hours. Mean with standard error are shown. L. Western blotting shows the nuclear TBX2 in a subtype I PDF upon TGF-β1 treatment and in a subtype III PDF upon TGFBR1i treatment. Histone H3 is used as a loading control. M. HGF and FGF7 expression in subtypes I (red) and II (green) PDFs upon TBX2 knockdown (siRNA pool). N. HGF and FGF7 expression in subtype III (blue) PDFs upon ectopic expression of TBX2. (M-N), knockdown and overexpression are confirmed by western blotting (left) and qRT-PCR (right),. (F-H, K, M-N), Paired one-tailed t-test is used. * p < 0.05, ** p < 0.01. See also Figures S7 and Table S4.
