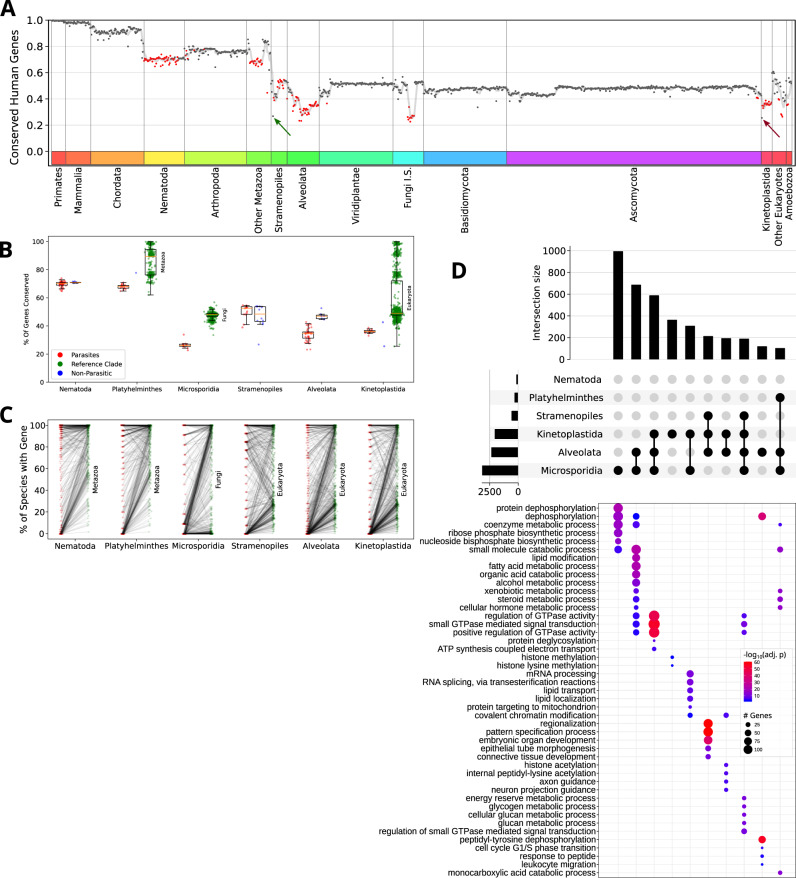
Fig. 7. Parasitic clades provide a key phylogenetic profiling signal.
The proportion of human genes found in each organism is shown on the y-axis, with parasites marked in red. Two non-parasitic organisms with lower conserved genes fraction are highlighted in green (Nannochloropsis gaditana, in Stramenopiles) and red (Perkinsela, in Kinetoplastida) arrows (A). The fraction of conserved genes was then compared for six clades with many parasitic organisms between parasites, non-parasitic organisms and a reference (parent) clade. Comparisons were made for each clade between parasitic organisms and the reference and non-parasitic organisms by a two-sided Mann–Whitney test; p-values are displayed for significant comparisons (p < 0.05). The boxplot extends from the lower to upper quartile values of the data, with an orange line at the median. Whiskers denote 1.5 times the interquartile range. (B). In addition to the species level, comparisons were made by pairing the average conservation for each gene in the parasitic organisms (red) and reference clade (green) with a line connecting them (C). Genes that were fully lost, or with low conservation in at least one parasitic clade but highly conserved across all Eukaryotes, were tested for losses combinatorics across these clades. Genes in the top 10 intersections were checked for gene ontology overrepresentation (biological process ontology). The top five terms by FDR adjusted p-value are shown for each combination (D). The upper panel presents the number of genes in each clade or intersection, with the relevant clades marked by black circles. The lower panels show the number and significance in the most relevant pathways. FDR—false discovery rate. Source data are provided as a Source Data file.