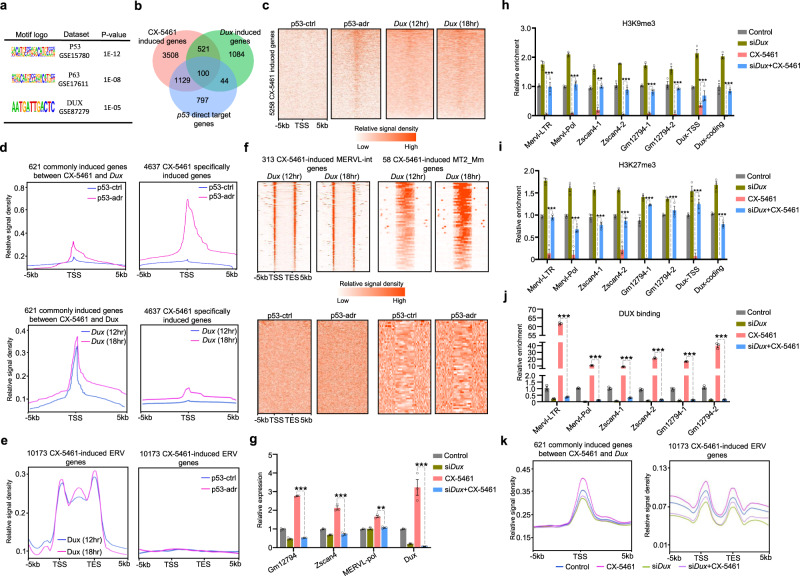
Fig. 5. 2C/ERV genes were activated through the Dux.
a Enriched binding motifs of 5258 genes induced by CX-5461 treatment. b Venn diagrams showing the overlap among CX-5461 treatment induced genes, p53 activated direct target genes and Dux-overexpression induced genes. c Heatmap plots demonstrate the binding of p53 and Dux proteins on within 5 kb region around transcription start sites of 5258 CX-5461 induced genes using published P53 ChIP-seq data and DUX ChIP-seq data. GEO accession code GSE26360 for P53 and GEO accession code GSE85632 for DUX . d Line plots demonstrate the meta-analysis results of P53 and DUX on within 5 kb region around transcription start sites of 621 commonly induced genes between CX-5461 treatment and Dux overexpression and 4637 specifically induced genes by CX-5461 using published P53 ChIP-seq data and DUX ChIP-seq data. GEO accession code GSE26360 for P53 and GEO accession code GSE85632 for DUX. e Line plots demonstrate the meta-analysis results of P53 and DUX protein on within 5kb region around transcription start and end sites of 10,173 CX-5461 induced ERV genes using published P53 ChIP-seq data and DUX ChIP-seq data. The regions of different lengths of gene body were fitted to 5 kb. GEO accession code GSE26360 for P53 and GEO accession code GSE85632 for DUX. f Heatmap plots demonstrate the binding of P53 and DUX proteins on within 5 kb region around transcription start and end sites of 5258 CX-5461 induced MERVL-int and MT2_Mm genes; The regions of different lengths of gene body were fitted to 5 kb. GEO accession code GSE26360 for P53 and GEO accession code GSE85632 for DUX. g qRT-PCR showing the expression of Dux or 2C-related genes in Dux silenced mES cells; GM12794: p = 1.21E−13(***), Zscan4: p = 3.00E−09(***), MERVL-pol: p = 4.55E−03(**), Dux: p = 9.60E−14(***), two-way ANOVA; N = 3 biologically independent ChIP-qPCR experiment; Data are presented as mean values +/− SEM. SEM standard error of mean. h ChIP-PCR showing H3K9me3 levels of Dux or 2C-related genes in Dux silenced mES cells; MERVL-LTR: p = 1.04E−11(***), MERVL-Pol: p = 9.68E−12(***), Zscan4-1: p = 2.51E−10(**), Zscan4-2: p = 1.67E−11(***), Gm12794-1: p = 1.90E−09(***), Gm12794-2: p = 2.66E−11(***), Dux-TSS: p = 1.35E−04(*), Dux-coding: p = 4.92E−11(***), two-way ANOVA; N = 3 biologically independent ChIP-qPCR experiment; Data are presented as mean values +/− SEM. SEM standard error of mean. i ChIP-PCR showing H3K27me3 levels of Dux or 2C-related genes in Dux silenced mES cells; MERVL-LTR: p = 9.87E−12(***), MERVL-Pol: p = 2.05E−08(***), Zscan4-1: p = 1.12E−11(***), Zscan4-2: p = 5.81E−10(**), Gm12794-1: p = 9.56E−12(***), Gm12794-2: p = 9.55E−12(***), Dux-TSS: p = 9.55E−12(***), Dux-coding: p = 1.02E−11(***), two-way ANOVA; N = 3 biologically independent ChIP-qPCR experiment; Data are presented as mean values +/− SEM. SEM standard error of mean. j ChIP-PCR showing DUX protein binding levels on 2C-related genes in Dux silenced mES cells; MERVL-LTR: p < 1.00E−15(***), MERVL-Pol: p < 1.00E−15 (***), Zscan4-1: p < 1.00E−15(***), Zscan4-2: p < 1.00E−15(***), Gm12794-1: p < 1.00E−15(***), Gm12794-2: p < 1.00E−15(***), two-way ANOVA; N = 3 biologically independent ChIP-qPCR experiment; Data are presented as mean values +/− SEM. SEM standard error of mean. k Line plots demonstrate the meta-analysis results of chromatin accessibility in Dux silenced mES cells within 5 kb region around transcription start sites or transcription start and end sites of 621 commonly induced genes between CX-5461 treatment and Dux overexpression and 10,173 CX-5461 induced ERV genes. The regions of different lengths of ERV genes were fitted to 5 kb. GEO accession code GSE166041. p53-ctrl: untreated mES cells, p53-adr: mES cells treated with adriamycin, a DNA damage agent widely used to activate p53, Dux (12 h): mES induced with doxycycline for 12 h, Dux (18 h): mES induced with doxycycline for 18 h. Source data are provided as a Source Data file.