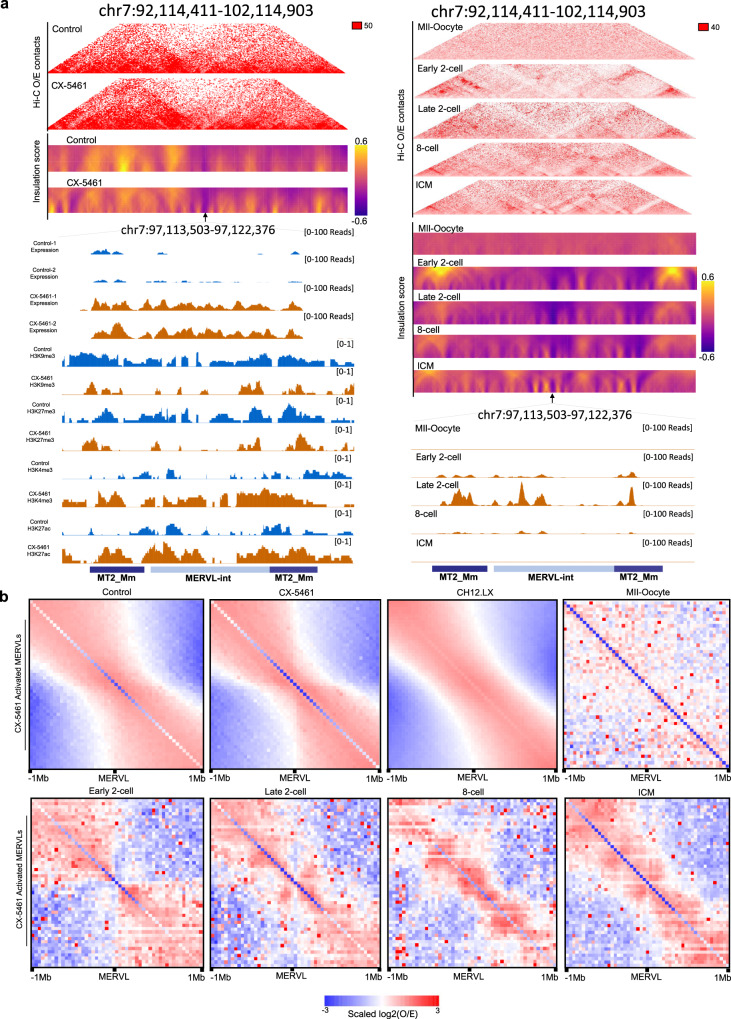
Fig. 7. rRNA biogenesis defect drove 3D chromatin structure reorganization of MERVL region towards the 2C-like state.
a Aggregate Observed(O)/Expected(E) Hi-C matrices centered on CX-5461 induced MERVL genes in control, CX-5461 treated mES cells and mouse embryos throughout mouse pre-implantation embryonic development. GEO accession codes GSE166041, GSE63525, and GSE82185. b Representative 40 kb Hi-C O/E interaction matrices of a MERVL loci located at TAD boundaries (chr7:97,113,503–97,122,376) are shown as heatmaps, along with the insulation score and genome browser tracks of RNA-Seq, H3K9me3, H3K27me3, H3K4me3, and H3K27ac ChIP-Seq signals of the expanded genomic region containing the TAD boundary (arrows) in control and CX-5461 treated mES cells as well as in mouse early embryos. GEO accession code GSE166041 and GSE82185.