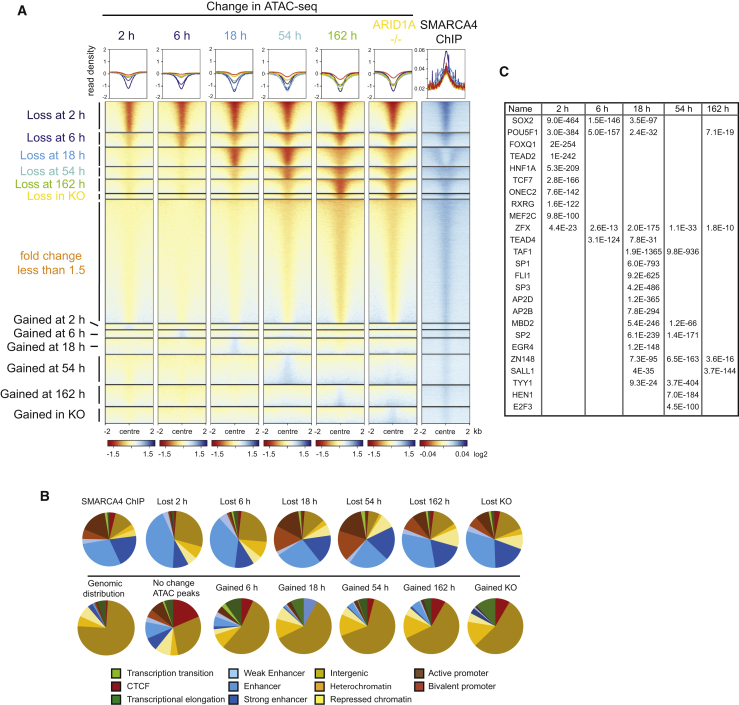
Figure 2.
Chromatin changes at different sites over time following ARID1A degradation
(A) Density plots of lost (red) and gained (blue) ATAC-seq signals changing at time points (2, 6, 18, 54, and 162 h) following degradation of ARID1A and in knockout (ARID1A−/−) mESCs. The list of sites is ordered by the time at which a change of >1.5-fold is first detected. (Last panel) Enrichment for ChIP-seq signal of the ATPase SMARCA4 (de Dieuleveult et al., 2016) at the same ATAC sites.
(B) Distribution of SMARCA4 ChIP-seq peaks and lost ATAC changes across ESC chromatin states (Pintacuda et al., 2017). Color code for chromatin states defined by histone modifications and transcription factor binding below the panel.
(C) Motif-based sequence analysis of ATAC peaks lost after 2, 6, 18, 54, and 162 h of ARID1A depletion. E values given show the significance of the motif as reported by MEME-ChIP (Motif Analysis of Large Nucleotide Datasets) (Machanick and Bailey, 2011). Only factors with an E value of 1 × 10 to 1 × 100 or lower are included.