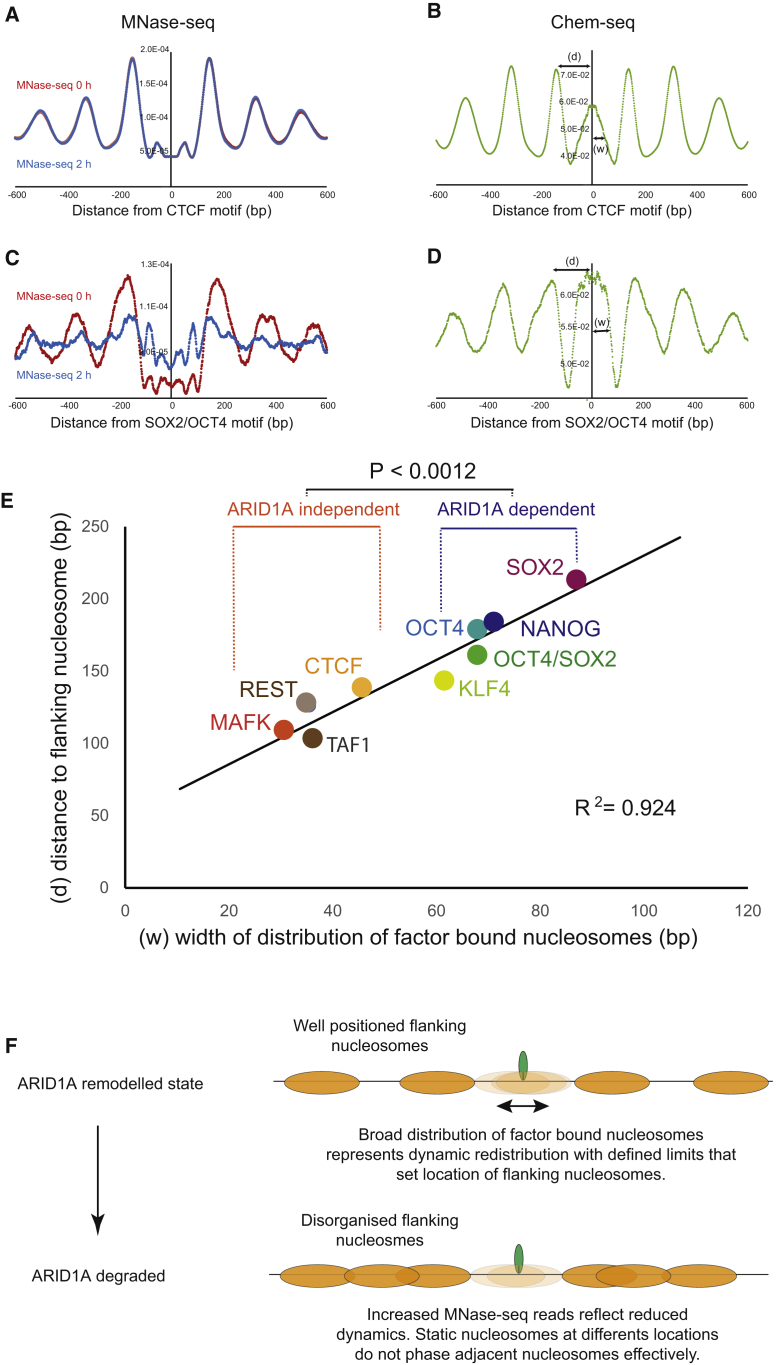
Figure 3.
ARID1A organizes clusters of nucleosomes adjacent to the binding sites of pluripotency transcription factors
(A) The centers of nucleosomal fragments protected from MNase digestion obtained prior to (red) and 2 h following degradation of ARID1A (blue) are shown aligned to the consensus binding site for CTCF.
(B) The distribution of chemical cleavage directed from histone H4 S47C (Voong et al., 2016) aligned to CTCF sites is shown in green. The distance to the adjacent nucleosome (d) and the width of the distribution of histone contacts (w) are indicated.
(C and D) Distributions of the same datasets at combined SOX2/OCT4 consensus motifs.
(E) Plot of the distance (d) between adjacent nucleosomes and the width of the distribution (w) at the DNA binding motifs for a range of transcription factors (see Figure S2).
(F) The limits of the distribution of ARID1A-sensitive transcription factor-bound nucleosomes have properties expected of a barrier capable of setting the locations of adjacent nucleosomes. In the absence of ARID1A a static distribution of nucleosomes across the factor binding site causes adjacent nucleosomes to be positioned heterogeneously.