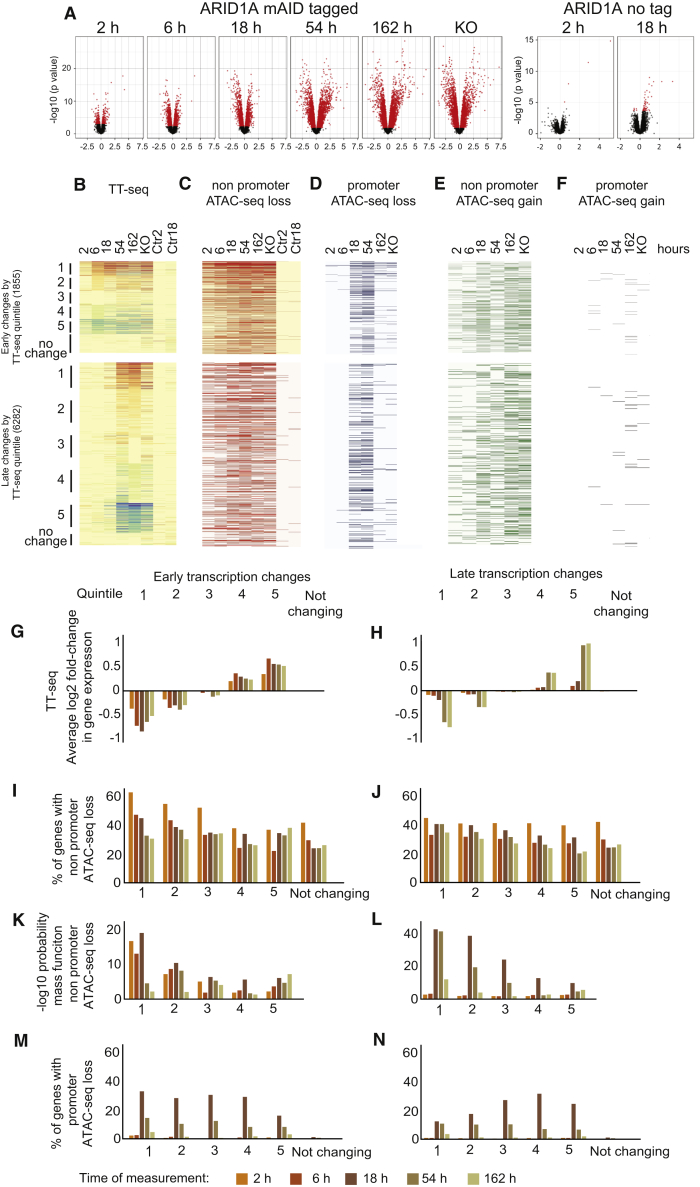
Figure 4.
Loss of enhancer chromatin accessibility is enriched at downregulated genes
(A) The numbers of genes changing significantly (red) at each time point represented as volcano plots. Right panels indicate changes following incubation of mESCs that do not have ARID1A mAID tagged with auxin.
(B) Transcription for genes changing (FDR < 0.05) after 2 and 6 h following ARID1A degradation and those changing later is plotted for groups sorted into quintiles based on the fold change at 6 and 162 h. A group of 500 genes selected based on no change to transcription following depletion of ARID1A is included for comparison. Changes in TT-seq at each time point are represented as a heatmap in (B). Also shown are the changes in an ARID1A −/− line and control lines in which ARID1A is not degron tagged. The average log2 fold change in expression for genes in each quintile at each time point is plotted as a histogram for early changing genes in (G) and for late genes in (H).
(C) Loss of ATAC-seq (> 1.5-fold) in the 50 kb region (excluding promoters) adjacent to differentially expressed genes.
(D) Loss of ATAC-seq (> 1.5-fold) at promoters (−500 to +500 bp from the TSS).
(E) Gain of ATAC-seq in regions 50 kb either side of the promoters of each TT-seq gene.
(F) Gained promoter ATAC-seq.
(G and H) Quintiles represent: 1, top 20% of repressed genes; 2, top 20%–40% of repressed genes; 3, top 40%–60% of genes; 4, top 20%–40% of activated genes; 5, top 20% of activated genes.
(I and J) Bar graphs showing the percentage of genes in each differentially expressed group of genes that have loss of chromatin accessibility within 50 kb on either side of the TSS. The different colored bars represent the percentage of genes that have sites of lost chromatin accessibility first detected at successive time points according to the key.
(K and L) −Log10 probability mass function for the enrichments in (I) and (J) are plotted indicating the likelihood that the enrichments would occur by chance.
(M and N) Percentages of genes in each group with promoter chromatin changes. In each histogram bars are colored according to the time at which changes in ATAC-seq are detected as indicated in the key.