Figure 5.
ARID1A-dependent EP300 occupancy in different chromatin contexts is associated with upregulation and downregulation of gene expression
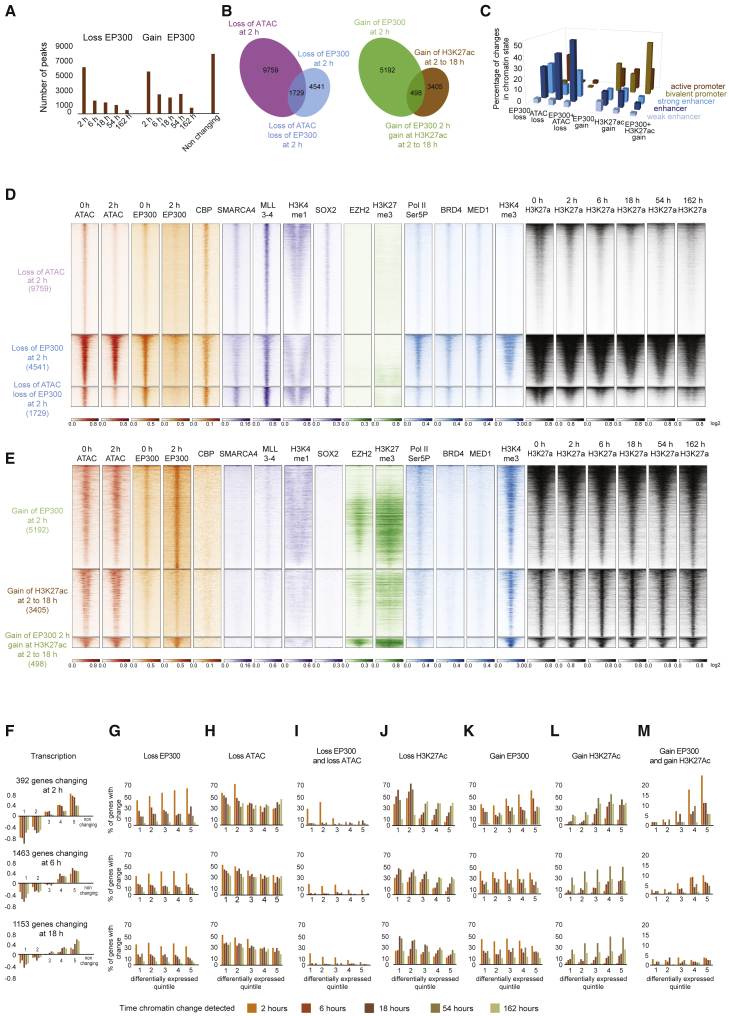
(A) The number of sites at which EP300 occupancy (FDR < 0.2 and fold change > 1.2) changes are detected are shown for each time point following degradation of ARID1A.
(B) Intersects between called changes in ATAC-seq, EP300 occupancy and histone acetylation.
(C) The distribution of the chromatin changes indicated in (B) across promoter and enhancer chromatin states (Pintacuda et al., 2017).
(D and E) Heatmaps showing enrichment for selected factors at the genes where chromatin changes indicated in (B) occur. Heatmaps are centered with 2 kb on either side.
(F) Average changes in transcription detected by TT-seq for quintiles of genes sorted by fold-change in expression and changing (FDR < 0.05) at 2, 6 and 18 h.
(G–M) The percentage of differentially expressed genes shown in (F) that have the indicated chromatin changes within a 50-kb flanking region that excludes the promoter (G–J). Sites of gained EP300 and histone acetylation are promoter proximal, so the proportion of genes with these changes is calculated including promoters (K–M). Data relating to the genes differentially expressed at 2, 6, and 18 h are shown in the top middle and bottom row of histograms. Each histogram is organized as a block of data for each differentially expressed quintile of genes that are first differentially expressed at the time point. For each quintile a different colored bar indicates the chromatin changes first detected at that time point. (M) Sites of histone acetylation accumulated during 2–18 h that coincide with gained EP300 occupancy at each time point.