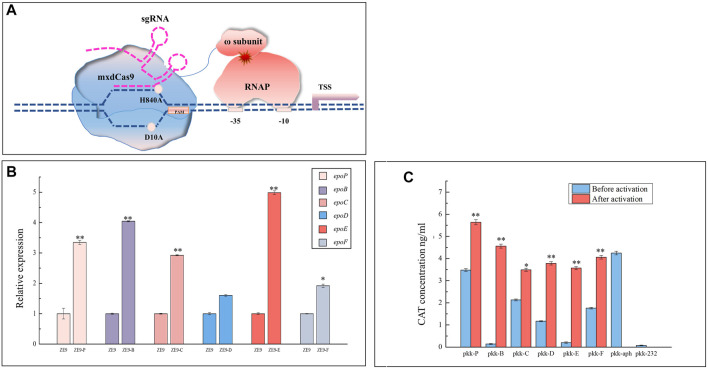
FIGURE 2.
Activation of internal promoters. (A) Principle of CIRSPR-dCas9 activation. mxdCas9: codon-optimized inactivated dCas9 protein (H840A, D10A); ɷ subunit: the Omega subunit of RNA polymerase; RNAP: RNA polymerase; sgRNA: single guide RNA sequence, a combination of the CRISPR-associated RNA (crRNA) and the trans-activation crRNA (tracrRNA). (B) RT-qPCR analysis of expression levels of the six operon genes activated by internal promoters in different mutants of M. xanthus ZE9 after 48 h of incubation. The gene expressions in M. xanthus ZE9 were each set as 1, and the expressions of the operon genes in mutant strains are shown as the relative expression. (C) Activities of the separate internal promoters in E. coli before and after activation by CRISPRa. The CAT activities were detected with the aphII promoter as a positive control and the original plasmid pKK-232 (no promoter upstream of the reporter gene CAT). The aphII promoter was not activated. The error bars in (B,C) represent the standard deviation of three independent experiments. For statistical analysis between the ancestral strain and mutant strains, the signals of ** and * mean p < 0.01 and p < 0.05, respectively.