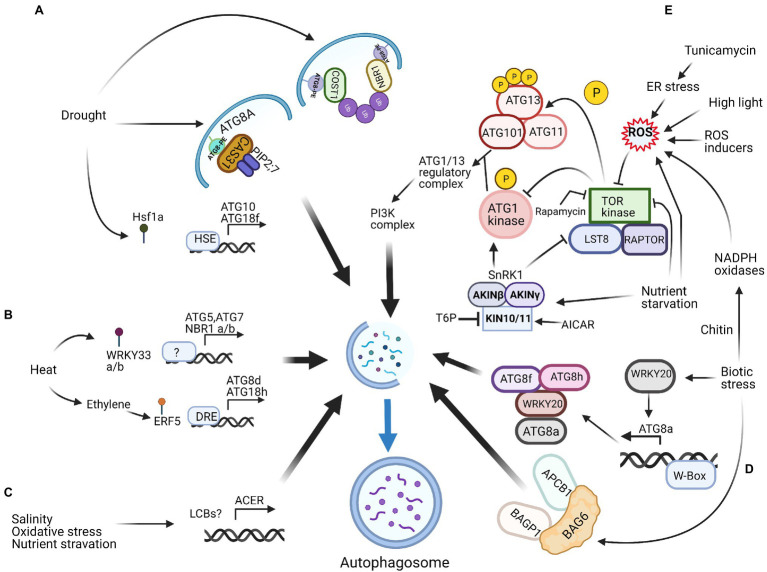
Figure 1.
Potential targets for manipulation of autophagy in plants. Identified regulators and potential ATG gene targets during abiotic stresses such as drought stress (A), heat stress (B), salinity, oxidative and nutrient starvation stresses (C), and biotic stresses (D) and stresses caused by reactive oxygen species (E). Refer text for the detailed information. NBR1, Neighbour of BRCA1; COST1, Constitutively Stressed 1; HSE, Heat shock element; ATG, Autophagy related; PE, Phosphatidylethanolamine; PIPs, Plasma membrane intrinsic proteins; Ub, Ubiquitin; HsfA1a, Heat-shock transcription factor A1a; ERF5, Ethylene response factor 5; DRE, Drought-responsive elements; ACER, Alkaline ceramidase; LCBs, Long‐chain bases; PI3K, Phosphatidylinositol-3-phosphate kinase; SnRK1, Sucrose nonfermenting1-related protein kinase1; TOR, Target of rapamycin; T6P, Trehalose-6-phosphate; AICAR, 5-Aminoimidazole-4-carboxamide ribonucleoside monophosphate; BAGP1, BAG-Associated GRAM Protein; APCB1, Aspartyl protease cleaving BAG; ROS, Reactive oxygen species. This figure was created with Biorender.