Abstract
To understand how the body of plants is made, it is essential to observe the morphology, structure and arrangement of constituent cells. However, the opaque nature of the plant body makes it difficult to observe the internal structures directly under a microscope. To overcome this problem, we developed a reagent, ClearSee, that makes plants transparent, allowing direct observation of the inside of a plant body without inflicting damage on it, e.g. through physical cutting. However, because ClearSee is not effective in making some plant species and tissues transparent, in this study, we further improved its composition to prevent oxidation, and have developed ClearSeeAlpha, which can be applied to a broader range of plant species and tissues. Sodium sulfite, one of the reductants, prevented brown pigmentation due to oxidation during clearing treatment. Using ClearSeeAlpha, we show that it is possible to obtain clear chrysanthemum leaves, tobacco and Torenia pistils and fertilized Arabidopsis thaliana fruits—tissues that have hitherto been challenging to clear. Moreover, we show that the fluorescence intensity of purified fluorescent proteins emitting light of various colors was unaffected in the ClearSeeAlpha solution; only the fluorescence intensity of TagRFP was reduced by about half. ClearSeeAlpha should be useful in the discovery and analysis of biological phenomena occurring deep inside the plant tissues.
Keywords: Clearing, Fluorescent microscopy, Fluorescent proteins, Oxidation, Reductant
Introduction
Biological processes occur inside and between cells in tissues. It is important to understand their roles and functions in vivo by analyzing the structure of the plant body. Many researchers have used microscopy to observe the inside of the body. Recently, several clearing reagents applicable to fluorescent microscopy using fluorescent proteins (FPs) in plants have been developed. Littlejohn et al. used perfluorocarbon to infiltrate the leaves and make them transparent to detect FPs (Littlejohn et al. 2010, Littlejohn and Love 2012, Littlejohn et al. 2014). In this reagent, leaves become clear in 5 min, but there is no reduction in the content of chlorophyll, which interferes with the visualization of FPs. Warner et al. (2014) performed clearing of plant tissues using Scale-based clearing reagents (urea, glycerol, Triton X-100). It took 1–3 weeks for leaves and root nodules to become transparent using these reagents. Hasegawa et al. (2016) and Musielak et al. (2016) used 2,2′-thiodiethanol to rapidly (1 h to 1 day) reduce the refractive index mismatch in plant tissues. These reagents can make plant tissues transparent, but the transparency is not sufficient to perform imaging of deep-seated structures in plants because they cannot remove chlorophyll (which causes autofluorescence). We also developed a clearing reagent, ClearSee, composed of urea, xylitol, and sodium deoxycholate, to perform whole tissue imaging using FPs, in tissues, such as roots, leaves, seedlings and pistils (Kurihara et al. 2015). Although this is a useful method for deep imaging of plants by removing chlorophyll, 3 d to 1 month are required to achieve transparency. ClearSee is applicable to a wide range of plant species, such as Arabidopsis thaliana, Physcomitrella patens (Kurihara et al. 2015), Astragalus sinicus (Ohtsu et al. 2017), avocado (Duman et al. 2020), barley (Ho et al. 2020), Brassica rapa (Arsovski 2020), Eucalyptus (Eliyahu et al. 2020), maize (Kelliher et al. 2017), Marchantia polymorpha (Aki et al. 2019), Monophyllaea glabra (Kinoshita et al. 2020), petunia (Chen et al. 2020), rice (Chu et al. 2018), soybean (Okuda et al. 2017), strawberry (Kim et al. 2019), Allium ochotense (Tanaka and Ono 2018), wheat (Wu et al. 2019) and Wolffiella hyalina (Isoda and Oyama 2018), it is not applicable to all tissues and species.
In this study, we found that oxidation causes brown pigmentation to form in the tissue during ClearSee treatment. Some reductants prevented the formation of this brown pigment while retaining the stability of the FPs used for imaging. We developed a modified clearing reagent, named ClearSeeAlpha, to prevent oxidation during clearing treatment. Moreover, we analyzed the stability of several FPs, emitting fluorescence from blue to red, in ClearSee solutions and found that TagRFP was not suitable for imaging using this clearing treatment. These findings demonstrate that ClearSeeAlpha allows deep imaging of a broad range of plant species and tissues using selected FPs.
Results and Discussion
ClearSee treatment induced proanthocyanidin derivative accumulation in some tissues
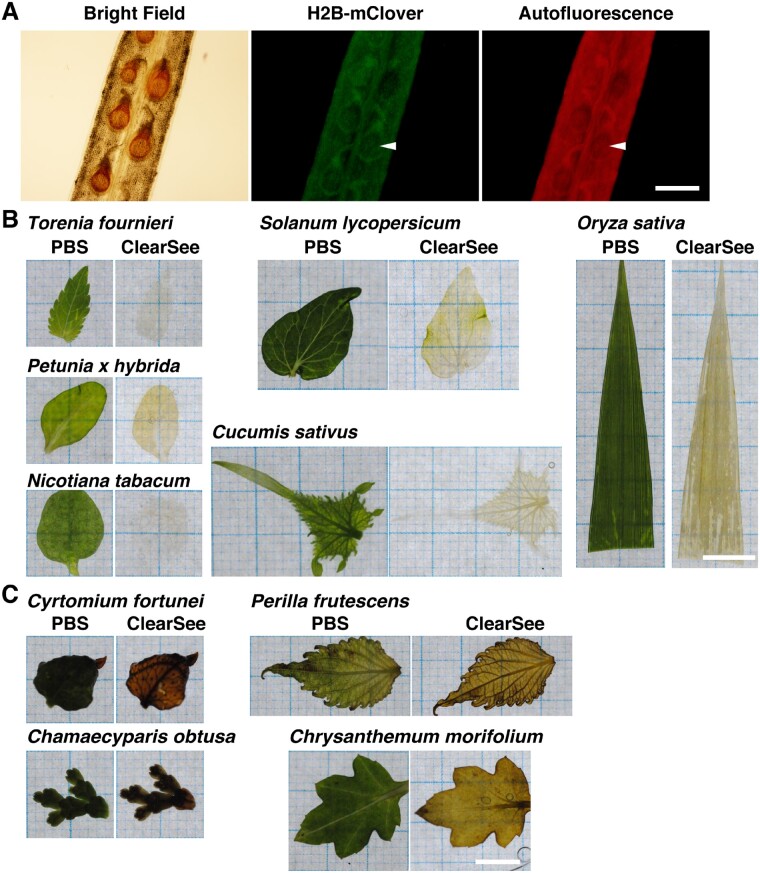
Previously, we developed a clearing reagent, ClearSee, to make transparent plant specimens for fluorescent microscopic observation (Kurihara et al. 2015). In A. thaliana, ClearSee could clear the roots, leaves, seedlings and pistils to maintain the stability of signals from FPs. However, we found that the ClearSee-treated siliques contained brown pigment on the seed coat (Fig. 1a, bright Field). The fluorescent signals were not detected in the regions of brown coloration, which instead appeared dark (Fig. 1A, H2B–mClover, Autofluorescence). Thus, this brown pigment interfered with the fluorescence imaging. The brown pigment on the seed coat was similar to that obtained with vanillin staining (Debeaujon et al. 2003). Vanillin staining can result in the visualization of proanthocyanidins as brown pigment owing to its oxidation. Proanthocyanidin derivatives accumulate on the seed coat during seed maturation after fertilization (Debeaujon et al. 2003). This brown pigment was not detected in the unfertilized ovules.
Fig. 1.
Optical clearing of tissues using ClearSee. (A) Fixed UBQ10p::H2B–mClover expressing pistil of A. thaliana was incubated in ClearSee for 6 d. Arrowheads indicate brown pigment regions in the bright field image. (B, C) Fixed leaves of various species were incubated in phosphate-buffered saline (PBS) or ClearSee for 8 d. The leaves were cleared (B) or not cleared in ClearSee. Scale bars: 500 µm (A) and 1 mm (B and C).
To identify plant species in which brown pigment appeared upon ClearSee treatment, we treated paraformaledehyde (PFA)-fixed leaves of different plants species with ClearSee. Like A. thaliana, leaves from most of the plants, including Torenia fournieri, Petunia x hybrida, Nicotiana tabacum, Solanum lycopersicum, Cucumis sativus and Oryza sativa, were cleared upon ClearSee treatment (Fig. 1B). Thus, various plant species, including horticultural plants and crops, both dicot and monocot, were compatible with ClearSee. However, ClearSee could not clear the leaves of Chamaecyparis obtusa and Cyrtomium fortunei (Fig. 1C). Because the leaves of plants, such as conifers and pteridophytes, are less permeable, the ClearSee solution could not penetrate inside the tissue. Moreover, leaves of Chrysanthemum morifolium and Perilla frutescens were not cleared even after 8 d of ClearSee treatment (Fig. 1C). The leaves retained green color in phosphate-buffered saline (PBS), whereas ClearSee-treated leaves displayed a brown pigment like in A. thaliana seeds. In tobacco, polyphenol oxidation by peroxidase causes brown pigmentation during leaf senescence (Sheen 1974). These results suggest that polyphenol, such as proanthocyanidin, oxidation induced the brown pigmentation observed in ClearSee-treated samples.
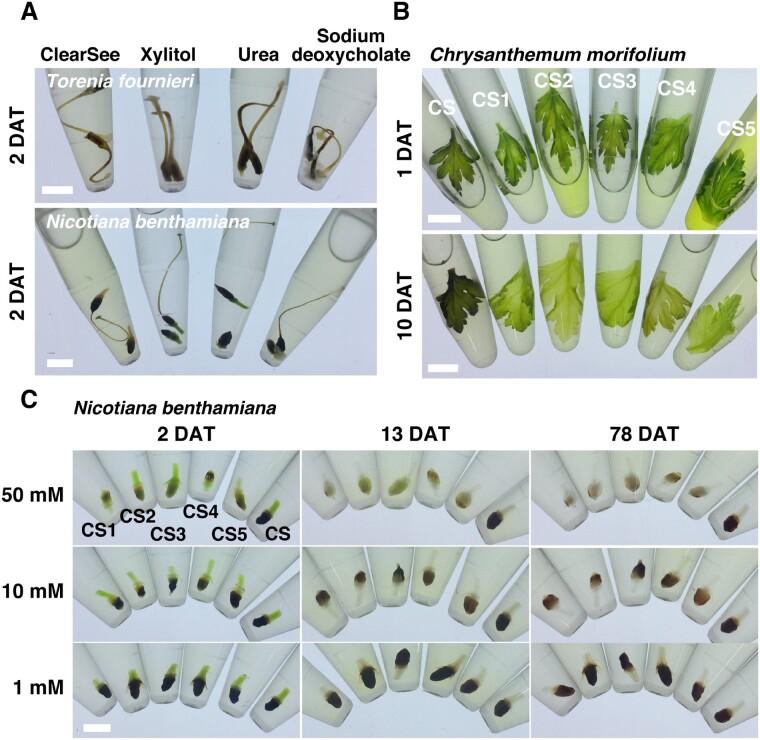
ClearSee consists of urea, xylitol and sodium deoxycholate (Kurihara et al. 2015). Torenia and tobacco pistils were very prone to brown pigmentation (Fig. 2A). To determine which of these ClearSee components are involved in oxidation, we incubated Torenia and tobacco pistils in urea, xylitol and sodium deoxycholate solutions. When incubated for 2 d, both Torenia and tobacco pistils turned brown in all the solutions (Fig. 2A). This indicates that oxidation occurred in each of the component of ClearSee. Moreover, precipitation was observed in the sodium deoxycholate solution (Fig. 2A). The absence of this precipitation in ClearSee suggests that sodium deoxycholate is homogenously dissolved in ClearSee.
Fig. 2.
Effect of reductants on clearing using ClearSee. (A) The pistils of T. fournieri and N. benthamiana were incubated in ClearSee, 10% (w/v) xylitol, 25% (w/v) urea, or 15% (w/v) sodium deoxycholate for 2 d. (B) The leaves of C. morifolium were incubated in modified ClearSee series for 1 and 10 d. (C) The ovaries of N. benthamiana were incubated in modified ClearSee series for 2, 13 and 78 d. Scale bars: 5 mm (A and C) and 1 cm (B).
Reductants prevent proanthocyanidin derivative accumulation in the leaf and ovary
Proanthocyanidins are colorless compounds, which are converted into brown derivatives by oxidation during seed maturation (Debeaujon et al. 2003, Pourcel et al. 2007). We speculated that if reductants suppress oxidation, it could prevent brown pigmentation. To search for reductants that prevent oxidation in ClearSee solution, we incubated chrysanthemum leaves and tobacco ovaries in ClearSee solution containing some reductants (Fig. 2B, C). We selected seven reductants commonly used in biochemical experiments for our analysis (Table 1). Reduced glutathione was rejected because of its insolubility; TCEP-HCl was rejected because of its high cost. Tobacco ovaries were incubated with various concentrations of 1-thioglycerol (CS1), 2-mercaptoethanol (CS2), DTT (CS3), 2-aminoethanethiol hydrochloride (CS4) and sodium sulfite (CS5) (Table 1). At 1 and 10 mM, brown pigment was seen 2 d after treatment (2 DAT) of leaves in all the reductant solutions. In tobacco ovaries incubated with 50 mM reductants, no brown pigment was observed at 2 DAT and transparency was seen at 13 DAT. At 78 DAT, there was no brown pigment in ClearSee containing reductants, and the transparency was increased.
Table 1.
List of reductants used for screening
| Reductant | Name | ¥/50 ml for 50 mM |
|---|---|---|
| 1-thioglycerol | CS1 | 147 |
| 2-mercaptoethanol | CS2 | 18 |
| DTT | CS3 | 879 |
| 2-aminoethanethiol hydrochloride | CS4 | 106 |
| Sodium sulfite | CS5 | 1 |
| Reduced glutathione | Not dissolved | 1,106 |
| TCEP-HCl | Expensive | 7,052 |
Similarly, when chrysanthemum leaves were incubated for 1 d, CS2 and CS5 solutions resulted in the elution of more chlorophyll in the solution; however, the leaves remained green (Fig. 2B; 1DAT). Further incubation resulted in the brown pigmentation of the leaves treated with ClearSee, but other leaves were pale green with reduced chlorophyll in CS1 to CS5 (Fig. 2B; 10DAT). These results indicate that each reductant’s addition can suppress the oxidation in ClearSee and prevent brown pigmentation of tissues.
Modified ClearSee stabilizes the FPs
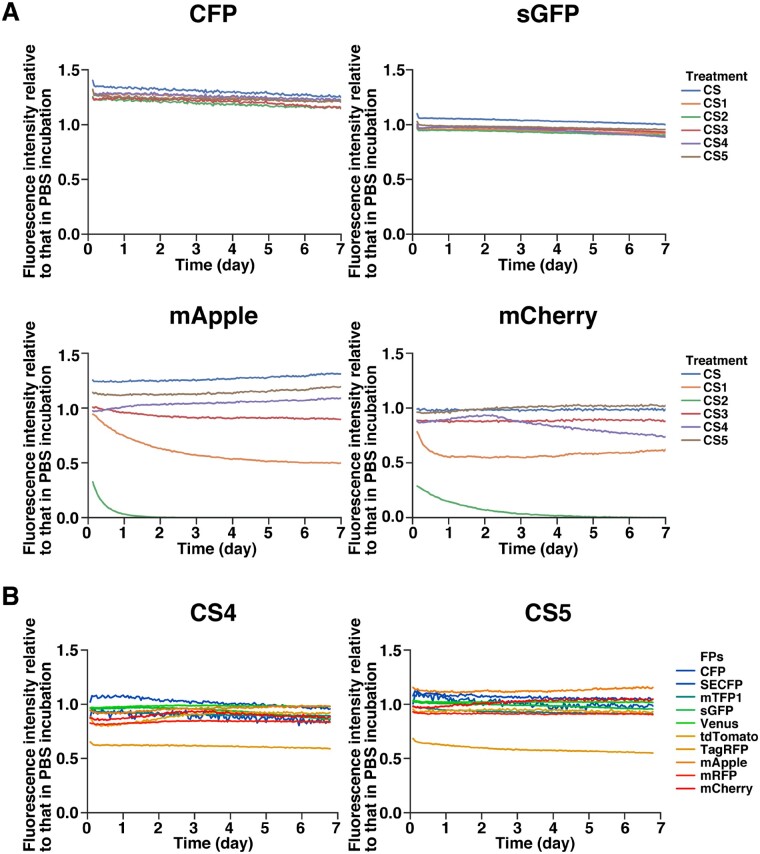
Next, to analyze the effect of reductants on the FPs, purified FPs were incubated in the modified ClearSee, and their fluorescence intensity was measured over time for 7 d (Fig. 3A). For CFP and sGFP, there was no significant change in fluorescence intensity in any of the modified ClearSee solutions (Fig. 3A; CFP, sGFP). By contrast, the fluorescence intensity of mApple and mCherry was decreased in some of the modified ClearSee solutions (Fig. 3A; mApple, mCherry). In CS2, the fluorescence was almost quenched after 1 d, and in CS1, the intensity was halved after 1–2 d. No significant differences were found for CS4 and CS5, while the fluorescence of mApple slightly decreased (Fig. 3A; mApple, mCherry). These results indicate that red FPs are affected by reducing agents depending on their type.
Fig. 3.
Stability of FPs in modified ClearSee series. Recombinant FPs were incubated with modified ClearSee series. The fluorescent signal intensities were measured every hour over 7-day incubation. Each plot shows the fluorescent intensities relative to that in the case of PBS incubation (mean, n = 3). (A) CFP, sGFP, mApple and mCherry were incubated in modified ClearSee series. The fluorescent intensities of mApple and mCherry were decreased in CS2 and CS3. (B) Different FPs were incubated in CS4 and CS5. The fluorescent intensity of TagRFP was decreased in CS4 and CS5.
To determine whether there is a difference in the effect of reductants on different types of FPs in more detail, the effects of CS4 and CS5 were analyzed (Fig. 3B). Among the 10 FPs that emitted fluorescence of different colors from blue to red, only TagRFP emitted fluorescence with half the intensity in both CS4 and CS5 (Fig. 3B). However, no decrease in fluorescence intensity was observed for other FPs, suggesting that CS4 and CS5 are useful for fluorescence imaging.
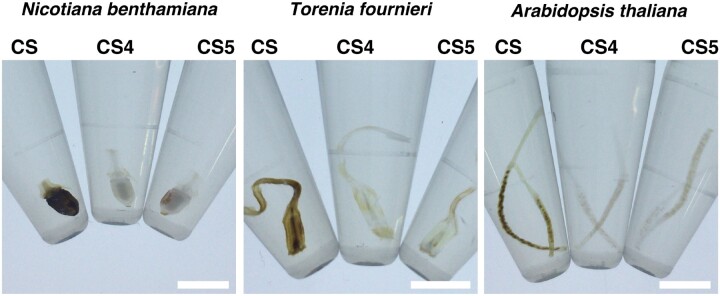
To determine whether there is a difference between CS4 and CS5, we treated Nicotiana benthamiana ovaries, N. tabacum pistils and A. thaliana siliques (Fig. 4). In ClearSee solution, all the ovaries showed brown pigmentation at 24 DAT. Although the surface of the ovary accumulated proanthocyanidin in N. benthamiana, the placenta or the seeds inside the ovary accumulated proanthocyanidin in Torenia, and A. thaliana, respectively (Fig. 4). In the case of CS4 and CS5, proanthocyanidin accumulation was inhibited in the treated samples (Fig. 4). There was no difference in transparency between the CS4 and CS5 treatments. The cost of sodium sulfate (CS5) compared with that of 2-aminoethanethiol hydrochloride (CS4) is 1/100; therefore, we preferred the use of CS5 as the new ClearSee reagent, which was named as ClearSeeAlpha.
Fig. 4.
ClearSeeAlpha prevents the formation of brown pigment during the clearing process. The ovaries of N. benthamiana, the pistils of T. fournieri and the siliques of A. thaliana were incubated in modified ClearSee series for 24 d. Scale bars: 5 mm.
ClearSeeAlpha enables deep imaging
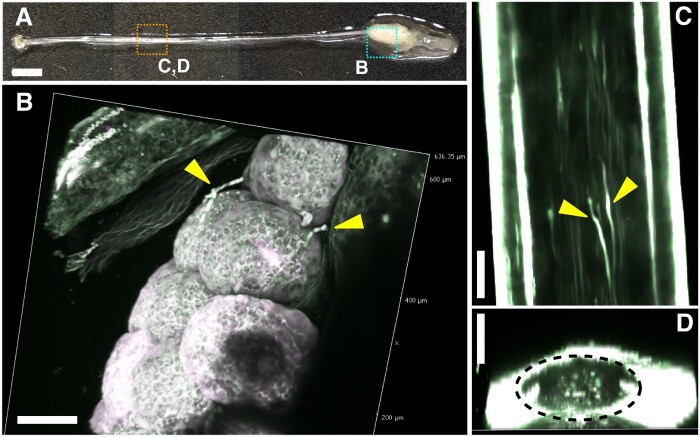
It is challenging to observe the pollen tubes elongating within the pistils of N. benthamiana. The style of N. benthamiana is very long and brittle, making it difficult to handle (Fig. 5A). Although the pollen tube elongation was observed with aniline blue staining (Isogai et al. 2020), it is still difficult to follow how the pollen tubes elongate within intact pistils without dissecting the style. Therefore, to confirm whether pollen tubes in intact pistils can be observed using ClearSeeAlpha, we prepared AtUBQ10pro::sGFP of N. benthamiana to label the pollen tubes. At 24 h after pollinating with AtUBQ10pro::sGFP pollen, the pistils were treated with ClearSeeAlpha for 3.5 months. Because N. benthamiana turns brown as soon as it is injured, we handled it gently. In the case of ovaries, ClearSeeAlpha could not make the ovary wall perfectly clear even after 3.5 months (Fig. 5A). We obtained the merged images from 31 z-stacks with 10-µm intervals using 800 nm excitation. As shown in Fig. 5B, we could detect the elongated pollen tubes on the ovules by removing the ovary wall. However, autofluorescence for green (519–549 nm) and red (560–630 nm) channels was also detected in the ovary (white signals in Fig. 5B). The elongated pollen tubes were detected without dissecting the style (Fig. 5C). The yz cross-section also showed elongated pollen tubes within the style (Fig. 5D).
Fig. 5.
Growth of a pollen tube in a pistil of N. benthamiana. (A) Nicotiana benthamiana pistil pollinated with UBQ10pro::sGFP pollen and treated with ClearSeeAlpha for 3.5 months. Cyan and orange dotted regions are magnified in (B) and (C) and (D), respectively. Reconstituted 3D images (B and D) or maximum-intensity projection for the xy view (C) were generated from 31 z-stack images with 10-µm intervals by 2PEM with 800 nm excitation. Yellow arrowheads indicate the elongated pollen tubes on the ovaries (B) or in the style (C). Yellow dotted region indicates the transmitting tract (D). Scale bars: 2 mm (A) and 100 µm (B)–(D).
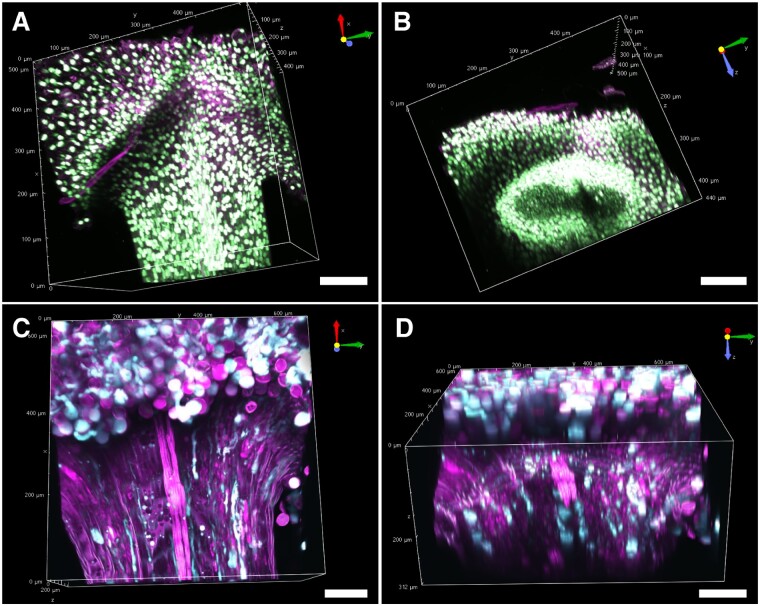
In the case of the stigma, the nuclei and the pollen tube were clearly detected in a ClearSeeAlpha-treated, AtUBQ10pro::H2B–mClover pistil (Fig. 6). We obtained the merged images from 88 z-stacks with 5-µm intervals using 920 nm excitation. As shown in Fig. 6A, we could detect the nuclei in the pollen grain, stigmatic papilla and style. The nuclei at the opposite side of the style from the objective lens were also detected in the ClearSeeAlpha-treated pistil (Fig. 6B). As shown in Fig. 6C, we were also able to detect the elongated pollen tube within the style in the ClearSeeAlpha-treated LAT52pro::mTFP1 pistil, in which the cytosol is labeled (Nagahara et al. 2021). We obtained the merged images from 26 z-stacks with 12-µm intervals using 930 nm excitation. In yz view, the position of the pollen tubes in the transmitting tract could be observed (Fig. 6D). The pollen tube position at the top of the style was broader than that of the middle of the style (Figs. 5D, 6D). Thus, ClearSeeAlpha is useful for observing the pollen tube elongation within pistils.
Fig. 6.
Cleared stigma of N. benthamiana. N. benthamiana stigma of UBQ10pro::H2B–mClover (A and B) and LAT52pro::mTFP1 (C and D) treated with ClearSeeAlpha for one month. Cyan shows the signal of mTFP1 (C and D), green shows that of H2B–mClover (A and B) and magenta shows autofluorescence signals at 560–630 nm (A–D).
Toward further whole-plant imaging
Because the style of N. benthamiana is easily broken and is difficult to use for semi-in vitro assay, we had not been able to observe the elongation of the pollen tube toward the ovule. We could observe the pollen tube elongation within the style and on the ovule in vivo by ClearSeeAlpha treatment. However, the ovary wall was not completely transparent and emitted red and green autofluorescence upon excitation at 800 nm; therefore, we should select the optimal FP for labeling pollen tubes. Two-photon excitation microscopy of orange FPs at the near-infrared wavelength (1,000 nm) enables us to reduce the autofluorescence and to detect the labeled target proteins in Arabidopsis tissues (Mizuta et al. 2015). TagRFP was not suitable for ClearSeeAlpha treatment (Fig. 3B). Other orange FPs, such as tdTomato and mApple, should be good candidates to label the protein of interest.
In Torenia, ClearSeeAlpha could make the whole flower transparent. As shown in Supplementary Fig. S1, the petals and sepals were cleared after 3 months of ClearSeeAlpha treatment. The ovary remained slightly colored, but most of the floral tissues were transparent. Although it takes a long time for the clearing treatment, ClearSeeAlpha was compatible for the clearing of large samples.
In this study, we developed a modified clearing solution, ClearSeeAlpha, to reduce the formation of brown pigment by preventing oxidation. The FPs in the ClearSee-treated samples are stable for more than a month (Kurihara et al. 2015). However, because ClearSeeAlpha has a reducing agent, it is necessary to pay attention to the following points:
Reducing agent activityBecause the reducing agent is easily deactivated, it is better to change the ClearSeeAlpha solution every week. ClearSeeAlpha can be made quickly by adding a reducing agent to ClearSee.
The quality of sodium deoxycholate. One notable thing is that the performance of clearing with ClearSee and ClearSeeAlpha depends on the quality of sodium deoxycholate. Sodium deoxycholate purchased from some manufacturers has a pale yellow color when dissolved and interferes with optical clearing and fluorescence imaging.
The time required for clearing with ClearSeeAlpha. Because reducing agents suppress brown pigment but do not accelerate chlorophyll removal, the time required for clearing with ClearSeeAlpha is the same as that with ClearSee. In A. thaliana, the required clearing times with ClearSee are 4 d for leaves and roots, 7 d for seedlings and 4 weeks for pistils (Kurihara et al. 2015). Tobacco and Torenia pistils also require 1–2 months to become transparent with ClearSeeAlpha. Note that larger tissues, such as Chrysanthemum leaves, require more time and more reagents (Fig. 2B).
Further optical clearing for whole-plant fluorescence imaging with ClearSeeAlpha will contribute to providing novel insights into the molecular mechanisms underlying various phenomena associated with plant development.
Materials and Methods
Plant materials and plant growth conditions
The leaves of the following plants were used for leaf clearing: C. obtusa, C. morifolium, C. sativus, C. fortunei, N. benthamiana, N. tabacum, P. x hybrida, P. frutescens and S. lycopersicum leaves were kindly provided by Dr. M. Notaguchi (Nagoya University). Oryza sativa leaf was kindly provided by Dr. R.D. Kasahara (Fujian Agriculture and Forestry University). We used A. thaliana accession Columbia and T. fournieri cv. ‘Blue and White’. The following transgenic lines were also used: UBQ10p::H2B–mClover of A. thaliana (Kurihara et al. 2015) and UBQ10p::sGFP, UBQ10p::H2B–mClover and LAT52pro::mTFP1 of N. benthamiana (Nagahara et al. 2021).
Arabidopsis thaliana seeds were sown on plates containing half-strength Murashige and Skoog salts (Duchefa Biochemie B.V., Haarlem, The Netherlands), 0.05% MES-KOH (pH 5.8), 1× Gamborg’s vitamin solution (Sigma, St. Louis, MO, USA) and 1% agar. The plates were incubated in a growth chamber at 22°C under continuous lighting after cold treatment at 4°C for 2–3 d. Two-week-old seedlings were transferred to the soil (Sakata no Tane; Sakata Seed, Yokohama, Japan) and grown at 21–25°C under long-day conditions (16-h light/8-h dark).
ClearSeeAlpha protocol
Tissue clearing was performed as described by Kurihara et al. (2015) with slightly modifications. ClearSee solutions were prepared by mixing xylitol powder [10% (w/v) final concentration; Wako, Osaka, Japan, 248-00545], sodium deoxycholate [15% (w/v) final concentration; Tokyo Chemical Industry, Tokyo, Japan, C0316] and urea [25% (w/v) final concentration; Wako, 211-01213] in water. The following reductants (50 mM final concentration) were used for screening: reduced glutathione (Wako, 071-02014); 1-thioglycerol (Nacalai Tesque, Kyoto, Japan, 33709-62); 2-mercaptoethanol (Wako, 131-14572); 1,4-dithiothreitol (Wako, 048-29224); 2-aminoethanethiol hydrochloride (Nacalai Tesque, 21419-32); and sodium sulfite (Wako, 190-03411). The leaves were fixed in 4% (w/v) PFA (Wako, 162-16065) for 120 min in PBS under vacuum (690 mmHg) at room temperature. Fixed tissues were washed twice for 1 min in PBS and cleared with modified ClearSee at room temperature or 4°C until clearing.
Fluorescence stability
The fluorescence stability of FPs in modified ClearSee was measured with a microplate reader (EnSpire; PerkinElmer, Waltham, MA, USA) as described previously (Kurihara et al. 2015). To prepare the recombinant FPs, the full-length coding regions of FPs (CFP, SECFP, mTFP1, sGFP, Venus, tdTomato, TagRFP, mApple, mRFP, mCherry) were cloned into the pCold I expression vector (Takara, Kyoto, Japan). The recombinant proteins were expressed in Escherichia coli strain Rosetta-gami2 (DE3) pLysS (Novegen, Darmstadt, Germany). After induction with 1 mM isopropyl-β-d-thiogalactopyranoside at 15°C overnight, cells were harvested and lysed in 20 mM phosphate buffer (pH 7.3) containing 500 mM NaCl (Wako, 191-01665), 5 mM imidazole (Wako, 097-05391), 1 mM 2-mercaptoethanol, 0.5% Tween-20 (Tokyo Chemical Industry, T0543), DNase (TURBO DNase, Invitrogen, Carlsbad, CA, USA) and cOmplete Protease Inhibitor Cocktail (Roche, Mannheim, Germany). After sonication and centrifugation, the supernatants were collected and applied on TALON resin (Clontech, Mountain View, CA, USA). After washing with 20 mM phosphate buffer (pH 7.3) containing 500 mM NaCl, 5 mM imidazole, the recombinant proteins were eluted with 20 mM phosphate buffer (pH 7.3) containing 500 mM NaCl, 50 mM imidazole and 1 mM 2-mercaptoethanol. Purified FPs were incubated in modified ClearSee solutions for 7 d, and the fluorescence intensities were measured at excitation and emission wavelengths of 485 and 515 nm, respectively, at every hour.
Microscopy settings
For deep imaging, we used a laser scanning inverted microscope (A1R MP; Nikon, Tokyo, Japan) equipped with a Ti:sapphire femtosecond pulse laser (Mai Tai DeepSee; Spectra-Physics, Mountain View, CA, USA). The Z-stack images were acquired using a 25× water-immersion objective lens (CFI Apo LWD 25× WI, numerical aperture = 1.10, working distance = 2.00 mm; Nikon). A Ti:sapphire laser was used for the excitation of mTFP1, sGFP and mClover. Fluorescence signals were detected by the external non-descanned GaAsP PMT detectors. We used dichroic mirrors, DM495LP and DM560LP, and the following band-pass filters, 479/40 nm for mTFP1, 534/30 nm for mClover and sGFP and 578/105 nm for autofluorescence. Images were processed with the NIS-Elements AR software 5.20.02 (Nikon) to create maximum-intensity projection images and to add color.
Funding
Japan Society for the Promotion of Science [Grant-in-Aid for Scientific Research on Innovative Areas (JP16H06464 to T.H.); Grant-in-Aid for Scientific Research (B, JP17H03697 to D.K.); Grant-in-Aid for Challenging Exploratory Research (JP18K19331 to D.K.); Grant-in-Aid for Scientific Research on Innovative Areas (JP20H05358 to D.K.); Grant-in-Aid for Transformative Research Areas (JP20H05779 to Y.M.)]; and the Japan Science and Technology Agency [PRESTO program (JPMJPR15QC to Y.M., JPMJPR18K4 to D.K.)]. The microscopy was partly supported by Live Imaging Center at the Institute of Transformative Bio-Molecules (WPI-ITbM) of Nagoya University.
Supplementary Material
Acknowledgments
We thank M. Notaguchi and R.D. Kasahara for the provision of materials, Y. Taniuchi, M. Suenaga and M. Fujii for assistance with preparing materials and Editage (www.editage.com) for English language editing.
Disclosures
No conflicts of interest are declared.
Contributor Information
Daisuke Kurihara, JST, PRESTO, Furo-cho, Chikusa-ku, Nagoya, Aichi, 464-8601 Japan; Institute of Transformative Bio-Molecules (ITbM), Nagoya University, Furo-cho, Chikusa-ku, Nagoya, Aichi, 464-8601 Japan.
Yoko Mizuta, Institute of Transformative Bio-Molecules (ITbM), Nagoya University, Furo-cho, Chikusa-ku, Nagoya, Aichi, 464-8601 Japan; Institute for Advanced Research (IAR), Nagoya University, Furo-cho, Chikusa-ku, Nagoya, Aichi, 464-8601 Japan.
Shiori Nagahara, Institute of Transformative Bio-Molecules (ITbM), Nagoya University, Furo-cho, Chikusa-ku, Nagoya, Aichi, 464-8601 Japan.
Tetsuya Higashiyama, Institute of Transformative Bio-Molecules (ITbM), Nagoya University, Furo-cho, Chikusa-ku, Nagoya, Aichi, 464-8601 Japan; Division of Biological Science, Graduate School of Science, Nagoya University, Nagoya, Aichi, 464-8602 Japan; Department of Biological Sciences, Graduate School of Science, University of Tokyo, 7-3-1 Hongo, Bukyo-ku, Tokyo, 113-0033 Japan.
References
- Aki S.S., Mikami T., Naramoto S., Nishihama R., Ishizaki K., Kojima M., et al. (2019) Cytokinin signaling is essential for organ formation in Marchantia polymorpha. Plant Cell Physiol. 60: 1842–1854. [DOI] [PubMed] [Google Scholar]
- Arsovski A.A. (2020). BrphyB is critical for rapid recovery to darkness in mature Brassica rapa. bioRxiv, 2020.05.22.111245.
- Chen M., Bruisson S., Bapaume L., Darbon G., Glauser G., Schorderet M., et al. (2020). VAPYRIN attenuates defence by repressing PR gene induction and localized lignin accumulation during arbuscular mycorrhizal symbiosis of Petunia hybrida. New Phytol., 229: 3481–3496. [DOI] [PMC free article] [PubMed]
- Chu T.T.H., Hoang T.G., Trinh D.C., Bureau C., Meynard D., Vernet A., et al. (2018) Sub-cellular markers highlight intracellular dynamics of membrane proteins in response to abiotic treatments in rice. Rice 11: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I., Nesi N., Perez P., Devic M., Grandjean O., Caboche M., et al. (2003) Proanthocyanidin-accumulating cells in Arabidopsis Testa: regulation of differentiation and role in seed development. Plant Cell 15: 2514–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duman Z., Hadas-Brandwein G., Eliyahu A., Belausov E., Abu-Abied M., Yeselson Y., et al. (2020) Short de-etiolation increases the rooting of VC801 avocado rootstock. Plants 9: 1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliyahu A., Duman Z., Sherf S., Genin O., Cinnamon Y., Abu-Abied M., et al. (2020) Vegetative propagation of elite Eucalyptus clones as food source for honeybees (Apis mellifera); adventitious roots versus callus formation. Israel J. Plant Sci. 67: 83–97. [Google Scholar]
- Hasegawa J., Sakamoto Y., Nakagami S., Aida M., Sawa S., Matsunaga S. (2016) Three-dimensional imaging of plant organs using a simple and rapid transparency technique. Plant Cell Physiol. 57: 462–472. [DOI] [PubMed] [Google Scholar]
- Ho W.W.H., Hill C.B., Doblin M.S., Shelden M.C., van de Meene A., Rupasinghe T., et al. (2020) Integrative multi-omics analyses of barley rootzones under salinity stress reveal two distinctive salt tolerance mechanisms. Plant Commun. 1: 100031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isoda M., Oyama T. (2018) Use of a duckweed species, Wolffiella hyalina, for whole-plant observation of physiological behavior at the single-cell level. Plant Biotechnol. 35: 387–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isogai M., Miyoshi K., Watanabe M., Yoshikawa N. (2020) Characterization of horizontal transmission of blueberry latent spherical virus by pollen. Arch. Virol. 165: 2807–2815. [DOI] [PubMed] [Google Scholar]
- Kelliher T., Starr D., Richbourg L., Chintamanani S., Delzer B., Nuccio M.L., et al. (2017) MATRILINEAL, a sperm-specific phospholipase, triggers maize haploid induction. Nature 542: 105–109. [DOI] [PubMed] [Google Scholar]
- Kim D.-R., Cho G., Jeon C.-W., Weller D.M., Thomashow L.S., Paulitz T.C., et al. (2019) A mutualistic interaction between Streptomyces bacteria, strawberry plants and pollinating bees. Nat. Commun. 10: 4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita A., Koga H., Tsukaya H. (2020) Expression profiles of ANGUSTIFOLIA3 and SHOOT MERISTEMLESS, key genes for meristematic activity in a one-leaf plant Monophyllaea glabra, revealed by whole-mount in situ hybridization. Front. Plant Sci. 11: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara D., Mizuta Y., Sato Y., Higashiyama T. (2015) ClearSee: a rapid optical clearing reagent for whole-plant fluorescence imaging. Development 142: 4168–4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlejohn G.R., Gouveia J.D., Edner C., Smirnoff N., Love J. (2010) Perfluorodecalin enhances in vivo confocal microscopy resolution of Arabidopsis thaliana mesophyll. New Phytol. 186: 1018–1025. [DOI] [PubMed] [Google Scholar]
- Littlejohn G.R., Love J. (2012) A simple method for imaging arabidopsis leaves using perfluorodecalin as an infiltrative imaging medium. J. Vis. Exp. 2: 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlejohn G.R., Mansfield J.C., Christmas J.T., Witterick E., Fricker M.D., Grant M.R., et al. (2014) An update: improvements in imaging perfluorocarbon-mounted plant leaves with implications for studies of plant pathology, physiology, development and cell biology. Front. Plant Sci. 5: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuta Y., Kurihara D., Higashiyama T. (2015) Two-photon imaging with longer wavelength excitation in intact Arabidopsis tissues. Protoplasma 252: 1231–1240. [DOI] [PubMed] [Google Scholar]
- Musielak T.J., Slane D., Liebig C., Bayer M. (2016) A versatile optical clearing protocol for deep tissue imaging of fluorescent proteins in Arabidopsis thaliana. PLoS One. 11: e0161107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahara S., Higashiyama T., Mizuta Y. (2021) Detection of a biolistic delivery of fluorescent markers and CRISPR/Cas9 to the pollen tube. bioRxiv, 2021.02.15.431139. [DOI] [PMC free article] [PubMed]
- Ohtsu M., Sato Y., Kurihara D., Suzaki T., Kawaguchi M., Maruyama D., et al. (2017) Spatiotemporal deep imaging of syncytium induced by the soybean cyst nematode Heterodera glycines. Protoplasma 254: 2107–2115. [DOI] [PubMed] [Google Scholar]
- Okuda A., Matsusaki M., Masuda T., Urade R. (2017) Identification and characterization of GmPDIL7, a soybean ER membrane-bound protein disulfide isomerase family protein. FEBS J. 284: 414–428. [DOI] [PubMed] [Google Scholar]
- Pourcel L., Routaboul J., Cheynier V., Lepiniec L., Debeaujon I. (2007) Flavonoid oxidation in plants: from biochemical properties to physiological functions. Trends Plant Sci. 12: 29–36. [DOI] [PubMed] [Google Scholar]
- Sheen S.J. (1974) Polyphenol oxidation by leaf peroxidases in Nicotiana. Botanical Gazette 135: 155–161. [Google Scholar]
- Tanaka E., Ono Y. (2018) Whole-leaf fluorescence imaging to visualize in planta fungal structures of Victory onion leaf rust fungus, Uromyces japonicus, and its taxonomic evaluation. Mycoscience 59: 137–146. [Google Scholar]
- Warner C.A., Biedrzycki M.L., Jacobs S.S., Wisser R.J., Caplan J.L., Janine Sherrier D. (2014) An optical clearing technique for plant tissues allowing deep imaging and compatible with fluorescence microscopy. Plant Physiol. 166: 1684–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Mock H.P., Giehl R.F., Pitann B., Mühling K.H. (2019) Silicon decreases cadmium concentrations by modulating root endodermal suberin development in wheat plants. J. Hazard. Mater. 364: 581–590. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.