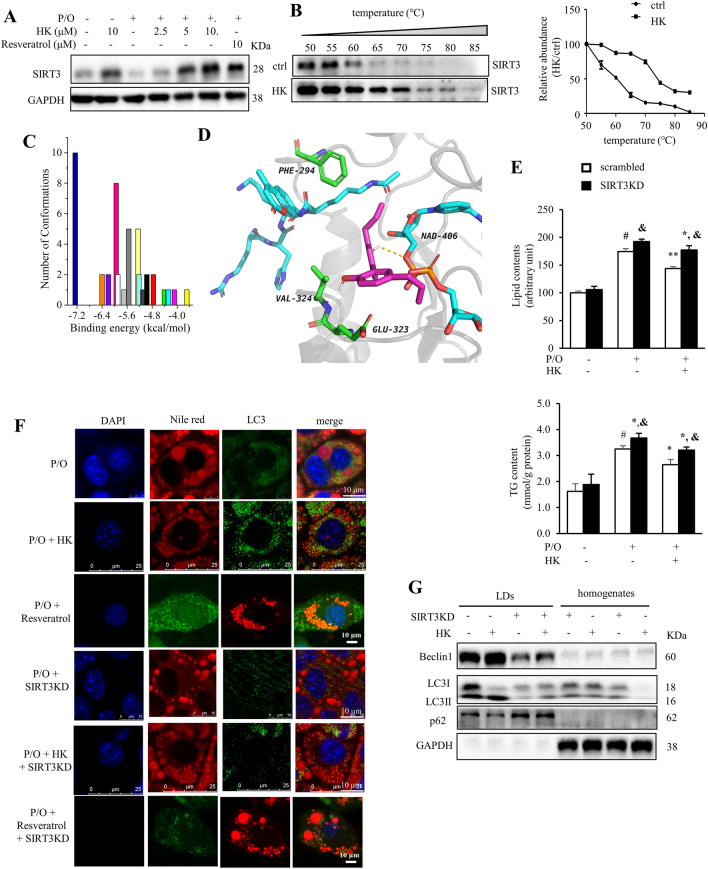
Fig. 2.
HK attenuates lipid accumulation through SIRT3-mediated autophagy. P/O-stimulated AML12 cells were treated without or with various concentrations of HK for 24 h. A SIRT3 protein levels were evaluated. GAPDH was used as a loading control. B CETSA was performed on AML12 cells treated with or without HK (10 μM) for 12 h. The SIRT3 protein levels were detected by using Western blotting. Data were normalized to the mean value of the respective group at 50 °C (n = 5). C Docking analysis of the binding between HK and SIRT3 (PDB ID: 5H4D). Cluster analysis of the docked conformations of HK. A tolerance of 2.0 Å was used. D Interactions between HK and residues on SIRT3. The protein was shown in New Cartoon, and small molecules in sticks; the substrate (or NAD+), residues, and HK were colored in cyan, green, and magenta, respectively. E The scrambled and SIRT3KD cells were treated with or without HK for 24 h. The lipid content was determined by flow cytometry after nile red staining and the cellular TG content were determined by commercial kit. F SIRT3 silencing blocked HK treatment-induced co-localization of LC3 puncta (green) on LDs. LDs were visualized with nile red fluorescence. Scale bar = 25 μm. G HK treatment activated autophagy mainly on LDs. Data was represented as means ± SD. *p < 0.05 and **p < 0.01, HK vs. P/O treatment. #p < 0.05, ctrl vs. P/O treatment. &p < 0.05, scrambled vs SIRT3KD groups. One-way ANOVA was used to calculate the p-values