The small GTPases Arf1–5 are important regulators of membrane traffic and Golgi functions. Pennauer et al. analyze viability and phenotypes of individual and combined CRISPR/Cas9 Arf knockout cell lines. Deletion of Arf1 or Arf4 produces specific phenotypes, while Arf4 alone is sufficient for cell viability.
Abstract
ADP-ribosylation factors (Arfs) are small GTPases regulating membrane traffic in the secretory pathway. They are closely related and appear to have overlapping functions, regulators, and effectors. The functional specificity of individual Arfs and the extent of redundancy are still largely unknown. We addressed these questions by CRISPR/Cas9-mediated genomic deletion of the human class I (Arf1/3) and class II (Arf4/5) Arfs, either individually or in combination. Most knockout cell lines were viable with slight growth defects only when lacking Arf1 or Arf4. However, Arf1+4 and Arf4+5 could not be deleted simultaneously. Class I Arfs are nonessential, and Arf4 alone is sufficient for viability. Upon Arf1 deletion, the Golgi was enlarged, and recruitment of vesicle coats decreased, confirming a major role of Arf1 in vesicle formation at the Golgi. Knockout of Arf4 caused secretion of ER-resident proteins, indicating specific defects in coatomer-dependent ER protein retrieval by KDEL receptors. The knockout cell lines will be useful tools to study other Arf-dependent processes.
Introduction
The secretory pathway is a major route of membrane traffic in the cell, transporting soluble and membrane proteins from their site of synthesis, which is the rough ER, to their final destinations. On the way, cargo proteins pass through successive compartments, where they acquire modifications and undergo multiple rounds of sorting and packaging into transport carriers. This anterograde traffic is counterbalanced by retrograde transport of membranes and proteins to maintain organelle identity and homeostasis, and retain specific proteins in defined compartments. Key players in these processes are the small GTPases of the ADP-ribosylation factor (Arf) family.
The Arf family is composed of 30 members: the six “true” Arfs, 21 Arf-like proteins (Arls), 2 Sars, and Trim23 (Li et al., 2004; Kahn et al., 2006). The Arfs are closely related, while the other members are more divergent in sequence and cellular functions (reviewed in Gillingham and Munro, 2007; Donaldson and Jackson, 2011; Sztul et al., 2019). The five human Arfs (humans lack Arf2) are assigned to three classes based on sequence homology: Arf1 and 3 belong to class I, Arf4 and 5 to class II, and Arf6 is the only member of class III. Class I and II Arfs mainly localize to the Golgi, but also to endosomes and/or the ER–Golgi intermediate compartment (ERGIC), whereas Arf6 is found in the cell periphery. Arfs are ubiquitously expressed, but vary in their abundance (Cavenagh et al., 1996; Itzhak et al., 2016). In the widely used HeLa cells, Arf1 is the most abundant Arf, followed by Arf4 (∼1/3), Arf5 and Arf6 (∼1/10), and Arf3 (∼1/100; Itzhak et al., 2016).
Arfs are N-myristoylated, which allows them to loosely associate with membranes already in the GDP-bound state. Binding of a guanine nucleotide exchange factor (GEF) and subsequent activation via GDP–GTP exchange lead to displacement of the N-terminal amphipathic helix from the hydrophobic binding pocket, resulting in tight membrane association (Antonny et al., 1997; Renault et al., 2003). Concomitant conformational changes enable binding of effectors.
The interplay of Arfs and their various effectors contributes to diverse cellular processes throughout the cell (reviewed in Donaldson and Jackson, 2011; Jackson and Bouvet, 2014; Sztul et al., 2019). The most prominent function is the contribution of the Golgi-localized Arfs (Arf1–5) to transport carrier formation in intracellular traffic, especially in the secretory pathway. Two major aspects are linked to Arf activity in this context: the modification of membrane lipids (reviewed in De Matteis and Godi, 2004; Donaldson and Jackson, 2011) and the recruitment of coat components. The best-characterized coat complexes are the coat protein complex I (COPI) at the ERGIC and the Golgi mainly for retrograde transport back to the ER, and the adaptor protein complex 1 (AP1) and the Golgi-localized, γ-ear-containing, Arf-binding proteins (GGAs) at the Golgi and on endosomes for transport from the TGN to endosomes and back.
The activity of Arfs is tightly regulated spatially and temporally by their GEFs and GTPase-activating proteins (GAPs). All 15 known GEFs share a common Sec7 domain to catalyze nucleotide exchange, but in addition possess diverse domains regulating their own membrane association and activity (reviewed in Nawrotek et al., 2016; Sztul et al., 2019). Also, the 28 ArfGAPs share a common GAP domain and are increasingly perceived to be more than simple terminators of Arf activity, but rather effectors themselves (Donaldson and Jackson, 2011; Sztul et al., 2019).
Originally discovered as a factor required for cholera toxin–mediated stimulation of adenylate cyclase by ADP-ribosylation of the stimulatory heterotrimeric G protein Gs (Kahn and Gilman, 1984, 1986), the role of Arfs in intracellular traffic by recruiting coat proteins was uncovered a few years later (Serafini et al., 1991; Stamnes and Rothman, 1993; Palmer et al., 1993; Traub et al., 1993). Early approaches to identify Arf functions were based on the manipulation by dominant-negative and dominant-active mutants (Teal et al., 1994; Zhang et al., 1994; Dascher and Balch, 1994) and by the fungal macrolide Brefeldin A. However, these approaches lacked specificity for individual Arfs, since Arf mutants and Brefeldin A sequester shared GEFs or GAPs and hence influence the activity of various Arfs simultaneously. Direct interactions between Arfs and coat components were analyzed in the presence of GTPγS by in vitro and in vivo experiments, which suggested that both class I and II Arfs can recruit COPI, AP1, and GGAs to Golgi membranes (Liang and Kornfeld, 1997; Boman et al., 2000; Austin et al., 2002; Takatsu et al., 2002).
Volpicelli-Daley and colleagues were the first to systematically dissect the role of individual Arfs in the secretory and endocytic pathway by siRNA-mediated knockdown. They proposed that no single Arf is required for any transport step, since only pairwise knockdowns resulted in specific phenotypes, hence suggesting cooperative action of Arfs and some isoform specificity at certain steps in intracellular transport (Volpicelli-Daley et al., 2005).
Later studies provided a glimpse into the complex interactome surrounding the Arfs involving GEFs, GAPs, effectors, and other GTPases, and highlighted that Arfs do not act in isolation, but in complex networks (reviewed in Donaldson and Jackson, 2011; Mizuno-Yamasaki et al., 2012; Baschieri and Farhan, 2012; Thomas and Fromme, 2016). However, fundamental questions remained unanswered, such as the major contributions of individual Arfs, their specificities and redundancies, and their regulation and coordination (Sztul et al., 2019).
Here, we revisited basic questions concerning the functions of Arf1–5 in the secretory pathway using CRISPR/Cas9 genome editing, generating Arf knockout (KO) cells by genomic deletion. We found that cells lacking any single Arf are viable, as well as cells deleted for certain double or triple combinations. In fact, Arf4 is able to sustain all essential functions in the absence of all other class I and class II Arfs. Yet we observed distinct phenotypes already in single-KO cell lines: deletion of Arf1 caused an increased Golgi volume, altered Golgi morphology, and reduced recruitment of vesicle coats to the Golgi, while the KO of Arf4 produced a specific defect in retrieval of ER resident proteins.
Results
Generation of Arf KO HeLaα cell lines
To characterize specific and redundant functions of Arf1–5 in the secretory pathway, we aimed to delete single or multiple Arf proteins by a CRISPR/Cas9-mediated KO. Two guide RNAs were designed to delete a genomic region of the respective Arf genes including the start codon and the part of exon 1 encoding the N-terminal myristoylated amphipathic helix.
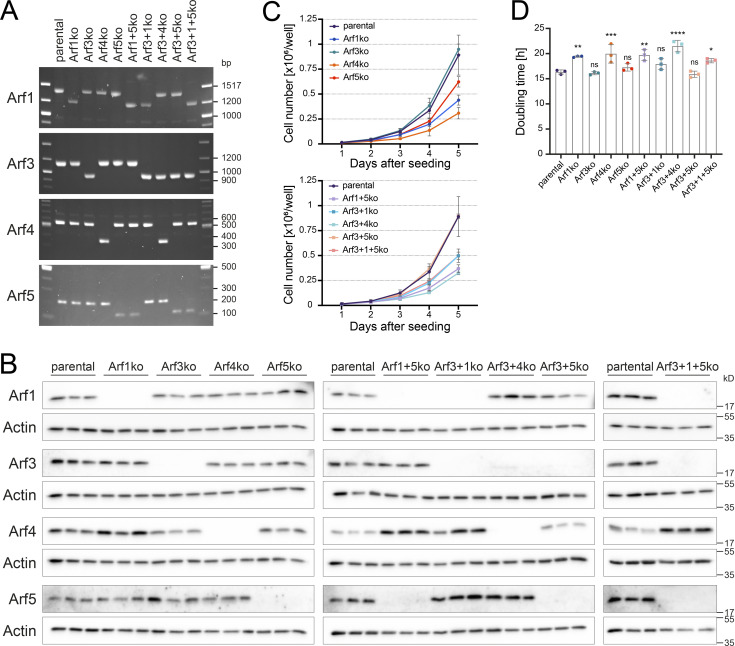
Initially, we knocked out single Arfs and succeeded to obtain all four KO cell lines (Arf1ko, Arf3ko, Arf4ko, and Arf5ko), indicating that no single Arf is essential for viability and cell growth (Table 1). Based on these single-KO cell lines, we generated cell lines for double- and triple-KO combinations. The chronological order of KOs is indicated in the name of the cell lines. In Arf3+1ko, for example, Arf3 was deleted first, followed by Arf1. Cell lines for four out of six double-KO combinations (Arf1+5ko, Arf3+1ko, Arf3+4ko, and Arf3+5ko) and one out of four triple-KO combinations (Arf3+1+5ko) were successfully generated, while we were repeatedly unable to obtain Arf1+4ko and Arf5+4ko cell lines (Table 1).
Table 1. Arf KO cell lines.
| KO | Single | Double | Triple |
|---|---|---|---|
| Deleted Arf(s) | 1 | 1+4 | 3+1+5 |
| 3 | 1+5 | ||
| 4 | 3+1 | ||
| 5 | 3+4 | ||
| 3+5 | |||
| 5+4 |
This overview lists all attempted Arf KO cell lines and indicates successful (highlighted in green) or failed generation (highlighted in red). The order of isoforms in double- and triple-KOs indicates the temporal succession of Arf deletion.
For all work presented here, one representative clonal cell line was carefully chosen for each KO after initial verification. This included validation of the deletions in all alleles by genomic PCR (Fig. 1 A) and confirmation of the KO on the protein level by immunoblot analysis (Fig. 1 B). In general, the loss of Arf proteins did not strongly affect the protein level of the remaining Arfs, with the exception of Arf4, which was reproducibly up-regulated in cells lacking Arf1 by an average of 4.1-fold (±0.9, mean ± SD; n = 3).
Figure 1.
Characterization of viable Arf KO cell lines. (A) Arf KO HeLaα cell lines were generated by CRISPR/Cas9 as described in Materials and methods and analyzed for partial deletion of exon 1 by genomic PCR. Uniform shortening of PCR fragments in KO cell lines indicated successful genomic deletion in all corresponding Arf alleles. (B) The deletion of Arfs on the protein level was verified by immunoblot analysis. For each cell line, three biological replicates were analyzed on the same gel. Molecular weight markers are indicated in kilodaltons. (C) Growth curves of Arf KO cell lines were derived from live cell counts obtained in three independent experiments (mean ± SD). Cells were seeded at the same density and monitored for 5 consecutive days. (D) Doubling times were calculated from the growth curves displayed in C. Bars, mean ± SD. Unpaired one-way ANOVA versus parental (ns, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
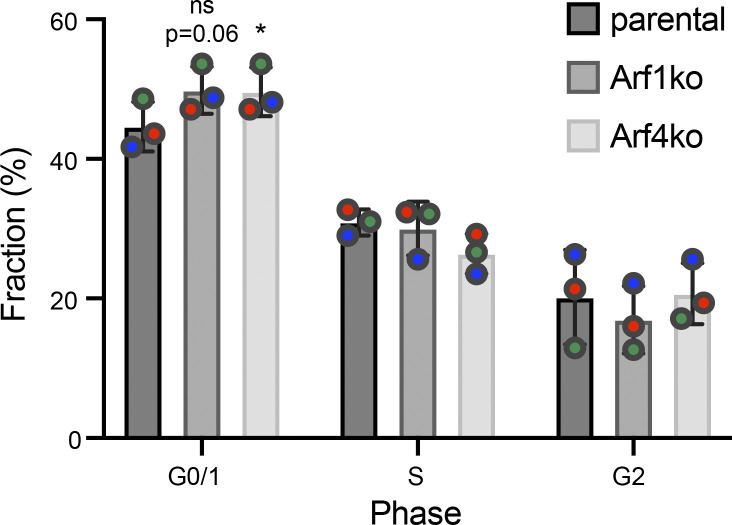
To test the impact of Arf deletions on cell growth, the doubling time was determined from growth curves based on live cell counts (Fig. 1, C and D). The growth rates of Arf3ko, Arf5ko, and Arf3+5ko cell lines were comparable to parental HeLaα cells. The other cell lines lacking Arf1 or Arf4, either alone or in combination with another Arf, grew more slowly, resulting in an increase in doubling time of ∼15% for cells without Arf1 and of ∼25% for Arf4-deleted cell lines. Consistent with the reduced growth rates, a larger fraction of Arf1ko and Arf4ko cells were in G0/1 phase of the cell cycle compared with parental HeLaα cells, as determined by DNA content (Fig. S1).
Figure S1.
Distribution of parental Helaα, Arf1ko and Arf4ko cells in the interphase. Cells were assigned to G0/1, S, or G2 phase based on their DNA content measured by Hoechst staining and flow cytometry (100,000 cells/experiment; n = 3). The fraction of cells in the respective phase is shown in percentage of the total. Mean ± SD of three experiments. Statistical significance was calculated for the G0/1 fraction, using paired one-way ANOVA versus parental (*, P < 0.05).
We conclude that no single Arf is required for cell survival. This is also true for several Arf KO combinations, with the exception of Arf4 in combination with Arf1 or Arf5. Simultaneous deletion of these Arfs appears to be lethal, since the respective KO cell lines could not be generated. Remarkably, Arf4 alone is sufficient for cell viability in the absence of all other class I and II Arfs. This highlights that class I Arfs are not essential.
Golgi morphology is altered and volume increased in cells deleted for specific Arfs
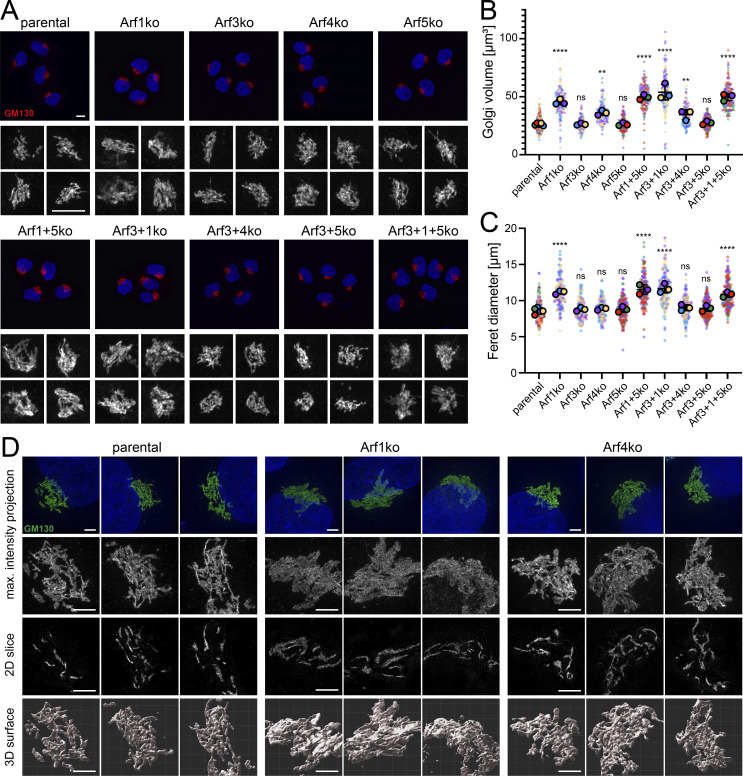
First, we assessed the impact of Arf deletions on the morphology of the Golgi complex by confocal microscopy acquiring serial z-stack images of cells immunostained for the cis-Golgi golgin GM130. In maximum intensity projections, the Golgi of parental HeLaα cells appeared as a perinuclear compact tangle of ribbons (Fig. 2 A). In all cell lines lacking Arf1 (Arf1ko, Arf1+5ko, Arf3+1ko, and Arf3+1+5ko), however, this shape was altered to a more diffuse and less compact pattern that appeared swollen and enlarged. Subtle changes were also suspected for cell lines lacking Arf4 (Arf4ko and Arf3+4ko), as the GM130-positive ribbons appeared to be more densely packed. No alteration was observed by eye in Golgi morphology of the other Arf KO cell lines.
Figure 2.
Impact of Arf KOs on Golgi morphology. (A) Maximum intensity projections were generated from serial confocal z-stack images of GM130-labeled Golgi. Lower panels show a magnified view of the Golgi complexes. Scale bars, 10 µm. (B and C) Volume and Feret diameter of individual Golgi complexes were determined using z-stack images described in A (on average 50 Golgi per cell line and experiment). Average values of three independent experiments are shown as filled circles together with the values from individual Golgi as small dots colored by experiment. Mean ± SD of the three experiments are indicated. Unpaired one-way ANOVA versus parental (ns, P > 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). (D) GM130 stained Golgi were imaged by super-resolution microscopy, and results are displayed as maximum intensity projections, tomographic 2D slices, and 3D reconstructed surfaces. DAPI is shown in blue. Lower panels show a magnified image section. Scale bars, 3 µm. max., maximum.
Golgi volume and Feret diameter (the largest distance between two contour voxels) were measured after 3D reconstruction and confirmed the visual evaluation (Fig. 2, B and C). All cell lines deleted for Arf1 displayed a significant increase in Golgi volume (on average 1.9-fold) and in the Feret diameter (on average 1.3-fold). In cell lines lacking Arf4, the Golgi volume was also significantly increased (on average 1.3-fold), whereas the Feret diameter remained at the levels of parental HeLaα cells. The other KO cell lines (Arf3ko, Arf5ko, and Arf3+5ko) did not exhibit a change in either parameter.
How these changes manifest themselves in the 3D structure of the Golgi was addressed by super-resolution microscopy (3D structured illumination microscopy [3D-SIM]; Fig. 2 D). In maximum intensity projections of 3D-SIM z-stacks, the GM130-labeled Golgi of parental HeLaα cells presented itself as a network of ribbons. In Arf1ko cells, this pattern was altered and appeared less defined and more uniformly distributed. Arf4ko Golgi displayed an intermediate phenotype. Tomographic 2D slices showed individual ribbons that were increased in the diameter in Arf1ko cells. In Arf4ko Golgi, the ribbons seemed to be more intertwined than in parental cells.
Golgi surface reconstruction showed a marked difference between a tangle of tubular structures in parental cells, clusters of more planar sheets in Arf1ko cells, and more densely packed Golgi ribbons in Arf4 KO cells. Taken together, the 3D analysis of the Golgi in KO cell lines confirmed an increase in Golgi volume for Arf1ko and Arf4ko cell lines and linked it to a broadening or higher number of Golgi ribbons, respectively.
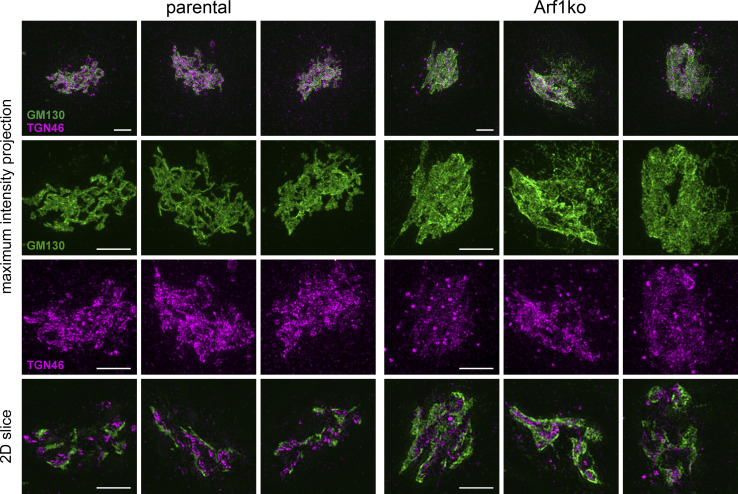
Maintenance of Golgi polarity in Arf1ko cells, which are the morphologically most affected KO cells, was assessed by super-resolution microscopy of coimmunostained cells. The separation of the cis-Golgi marker GM130 and the TGN marker TGN46 in Arf1ko Golgi complexes was comparable to parental cells, thus indicating that Golgi polarity is not perturbed (Fig. S2).
Figure S2.
Golgi polarity is maintained in Arf1ko cells. Golgi complexes from parental HeLaα and Arf1ko cells were immunostained for the cis-Golgi marker GM130 (green) and the TGN marker TGN46 (magenta). Images were acquired as confocal z-stack images using super-resolution microscopy and displayed as maximum intensity projections or tomographic 2D slices. The lower panels show magnified image sections. Scale bars, 3 µm.
Morphological changes in the ultrastructure of Arf1 KO cells
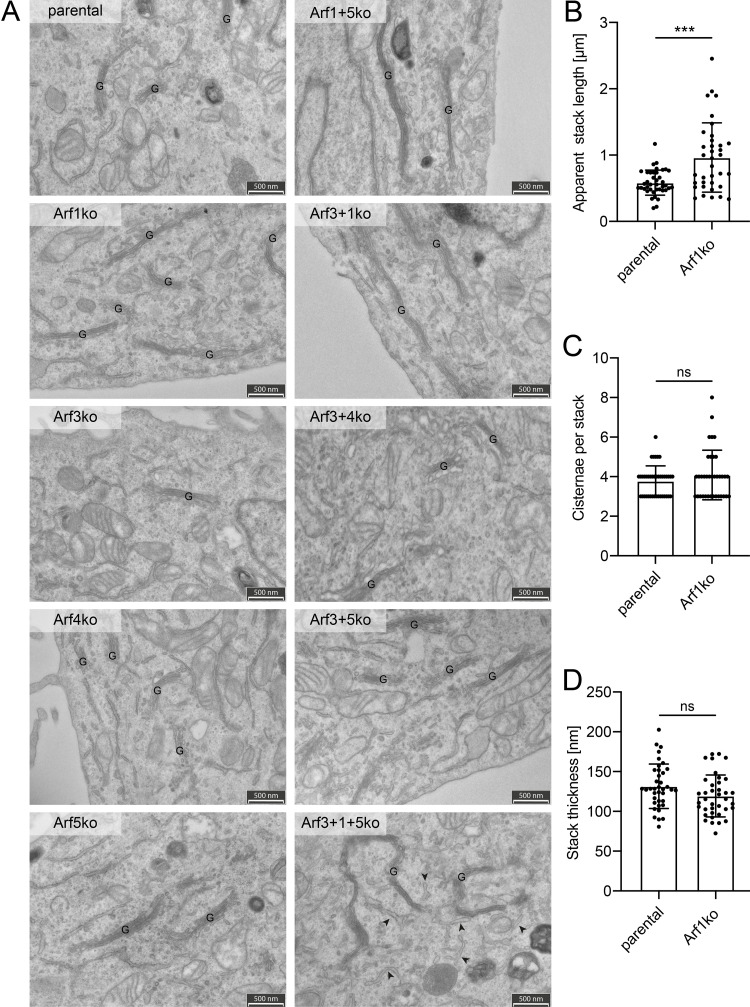
We further analyzed the ultrastructure of the Golgi by thin-section transmission electron microscopy (Fig. 3 A). The only difference in the appearance of Golgi structures was an increase in the length of individual stacks in cell lines lacking Arf1 compared with parental HeLaα cells. Quantitation revealed a significant increase in stack length of ∼65% in Arf1ko cells, while stack thickness and the number of cisternae per stack remained unchanged (Fig. 3, B–D). This is consistent with the increased ribbon diameter observed by super-resolution microscopy. No indication of morphological changes in Golgi structure was seen in other cell lines.
Figure 3.
Analysis of the ultrastructure of the Golgi. (A) Thin section transmission electron microscopy was performed to examine the ultrastructure of cellular compartments in all Arf KO cell lines. The field of view was chosen to include Golgi stacks in the acquired images. G, Golgi stack; arrowheads indicate dilated ER; scale bar, 500 nm. (B–D) Individual Golgi stacks were quantified for their apparent length (B), number of cisternae per stack (C), and thickness (D) in electron microscopy images of HeLaα and Arf1ko cells. Approximately 40 Golgi stacks were quantified per cell line. Bars, mean ± SD. Unpaired, two-tailed t test (ns, P > 0.05; ***, P < 0.001).
Interestingly, in the triple-KO cell line (Arf3+1+5ko), the tubular elements of the ER, as identified by bound ribosomes, appeared frequently dilated (Fig. 3 A, arrowheads).
KO of specific Arfs affects steady-state localization of coats
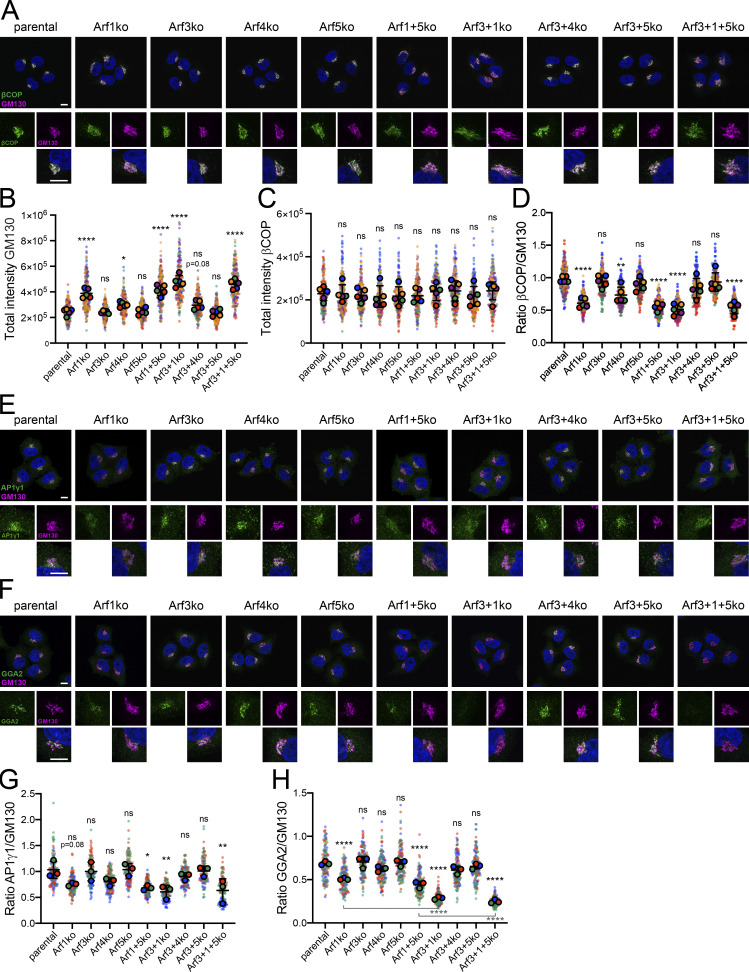
Next we examined the impact of Arf deletions on the recruitment of the Golgi-associated, Arf-dependent coats, COPI, AP1, and GGA2, by analyzing their steady-state localization by immunofluorescence microscopy in cells costained for the Golgi marker GM130.
The signal for βCOP mostly colocalized with GM130 (Fig. 4 A). However, in all Arf1 KO cell lines, the fluorescent βCOP signal appeared to be reduced at the Golgi compared with parental HeLaα cells. No change was observed in the other Arf KO cell lines.
Figure 4.
Immunostaining for Golgi associated coat proteins. (A) All Arf KO cell lines as well as parental HeLaα cells were coimmunostained for the Golgi marker GM130 in magenta and the coat component βCOP in green to examine their steady-state localization. DAPI signal is shown in blue. Lower panels show magnified image sections. Scale bars, 10 µm. (B–D) The total fluorescence intensity of perinuclear GM130 (B) and βCOP (C), and the ratio of βCOP to GM130, i.e., the density of βCOP per Golgi unit (D), were quantified. Average values obtained from five independent experiments with >40 Golgi analyzed per cell line and experiment are shown as filled circles together with the values from individual cells as small dots colored by experiment. Two of the five experiments were acquired later (violet and orange) and, due to technical offset, were mathematically corrected by a factor determined from the average ratios of the parental cells. Mean ± SD of the five experiments are indicated. Unpaired one-way ANOVA versus parental (ns, P > 0.05; *, P < 0.05; **, P < 0.01; ****, P < 0.0001). (E and F) The cell lines were stained for GM130 and AP1γ1 (E) or GGA2 (F) and imaged as in A. (G and H) The ratio of fluorescence intensity of AP1γ1 (G) or GGA2 (H) to GM130 was quantified as in D. Average values from three independent experiments with >40 Golgi analyzed per cell line and experiment are shown as filled circles together with the values from individual cells as small dots colored by experiment. Mean ± SD of the three experiments are indicated. Unpaired one-way ANOVA versus parental, unless indicated otherwise (ns, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
To substantiate this visual assessment, we quantified the signal intensity of βCOP and GM130 at the Golgi. Total fluorescence intensity revealed a significant increase of Golgi-localized GM130 in cells lacking Arf1 or Arf4 (Fig. 4 B), replicating the increase in Golgi volume described above. The total signal of βCOP, on the contrary, did not differ between cell lines (Fig. 4 C). As a consequence, the ratio of βCOP to GM130, i.e., the βCOP density at the Golgi, was reduced by an average of 40–50% in the Golgi region of all Arf1 KO cell lines compared with parental cells (Fig. 4 D). For cell lines lacking Arf4, a slight, although mostly not significant, decrease of Golgi-localized βCOP was observed. Since the total protein level of βCOP remained unchanged in Arf KO cells (Fig. S3), the results indicate reduced βCOP recruitment per Golgi unit.
Figure S3.
Intracellular βCOP levels. Levels of βCOP in parental HeLaα and Arf KO cell lines were analyzed by immunoblot analysis with actin as a loading control. For each cell line, three biological replicates were analyzed on the same gels with parental lysates.
The TGN coats AP1γ1 and GGA2 were analyzed following the same procedure using GM130 as a Golgi marker (Fig. 4, E and F) or TGN46 as a TGN marker (Fig. S4, A and B). As for βCOP, Golgi-localized AP1γ1 was reduced relative to GM130 by an average of 30–40% in all Arf1 KO cell lines compared with parental cells (Fig. 4 G). Similarly, the density of GGA2 at the Golgi was also reduced in all Arf1-deleted cell lines (Fig. 4 H). Remarkably, the additional absence of Arf3 in Arf3+1ko and Arf3+1+5ko cells even enhanced the loss of GGA2 at the Golgi compared with Arf1ko and Arf1+5ko cells from ∼30% to ∼60%. For cell lines lacking Arf4, a slight, although mostly not statistically significant, decrease in Golgi density of AP1γ1 and GGA2 was observed.
Figure S4.
Coimmunostaining for the TGN marker TGN46 and coat proteins. Parental HeLaα cells and all Arf KO cell lines were coimmunostained for TGN46 in magenta and AP1γ1 (A) or GGA2 (B) in green, respectively. DAPI signal is shown in blue. Lower panels show magnified image sections. Scale bars, 10 µm.
These findings suggest that normally, Arf1 is the main mediator of coat recruitment at the Golgi. In its absence, the rate of coat recruitment and thus of the formation of Golgi-exit carriers per Golgi unit is reduced, which results in expansion of the Golgi to match influx and efflux of proteins and membranes in a new steady-state.
KO of Arf4 causes defective retrieval of ER resident proteins
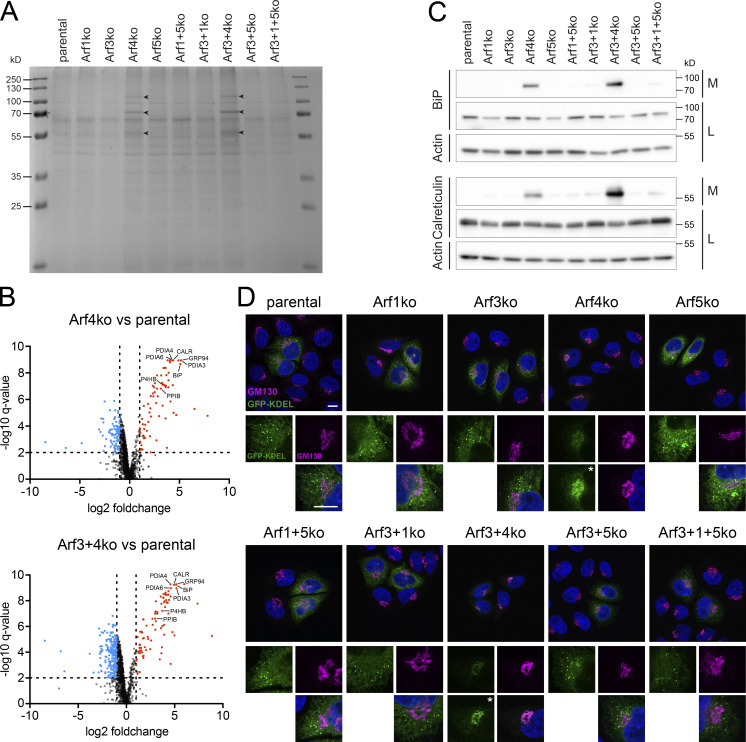
To assess the functionality of the secretory pathway upon deletion of Arfs, we analyzed total secretion by visualizing secreted proteins collected from the media by SDS-gel electrophoresis and Coomassie staining (Fig. 5 A). Strikingly, in the media of Arf4-deleted cell lines, additional bands were detected. For identification of these additionally secreted proteins, media were collected from Arf4ko, Arf3+4ko, and parental HeLaα cells and analyzed by mass spectrometry. This approach identified 75 and 87 proteins to be significantly up-regulated (as defined by fold change >2, q-value <0.01) in the secretomes of Arf4ko and Arf3+4ko cell lines, respectively (Fig. 5 B, Table S1, and Table S2). 70 of these were shared between the two KO cell lines. Among the top hits, we found ER chaperones, such as BiP, calreticulin, and GRP94, peptidyl-prolyl cis/trans isomerases, and protein disulfide isomerases. Gene ontology term (GOterm) enrichment analysis identified the ER as the main compartment of origin (Table S3 and Table S4). Secretion of BiP and calreticulin was verified by immunoblot analysis (Fig. 5 C). Both chaperones were found to be strongly secreted specifically in the two cell lines lacking Arf4.
Figure 5.
Examination of total secretion and KDEL-based retrieval of ER resident proteins. (A) Secreted proteins collected from the media of Arf KO cell lines were visualized on Coomassie stained SDS-gels. Arrowheads indicate additional bands. (B) Volcano plots of ∼1,400 secreted proteins, which were detected by mass spectrometry in the media of parental HeLaα, Arf4ko, and Arf3+4ko cells. Proteins enriched in KO versus parental samples were marked in red (fold change >2, q-value <0.01) and depleted ones in blue (fold change <0.5, q-value <0.01). Analysis was performed in biological quintuplicates. Dotted lines indicate the significance thresholds. Some ER proteins mentioned in the text are identified with their gene names. (C) Media collected from the indicated cell lines (M) and the corresponding cell lysates (L) were probed for the presence of the two ER chaperones BiP and calreticulin by immunoblotting. Actin served as a loading control. (D) Steady-state localization of transiently expressed signal sequence–GFP–KDEL (green) coimmunostained with GM130 (magenta). DAPI signal is shown in blue. Lower panels show magnified image sections. Asterisks mark images with enhanced intensity. Scale bars, 10 µm.
Aberrant secretion of ER resident proteins in Arf4-deleted cells indicates a defect in their retrieval from the Golgi back to the ER. The most prominent mechanism is retrograde transport by the KDEL receptors (Lewis and Pelham, 1990, 1992; Hsu et al., 1992; Raykhel et al., 2007). Indeed, ∼30% of the proteins whose secretion was increased in Arf4 KO cells contain a KDEL motif or a variant thereof at their C terminus according to the PROSITE database (https://prosite.expasy.org) consensus sequence [KRHQSA]-[DENQ]-E-L> (entry PDOC00014). However, this might be an underestimation, as studies suggested that not all motifs recognized by the KDEL receptors are included in this consensus pattern (Raykhel et al., 2007).
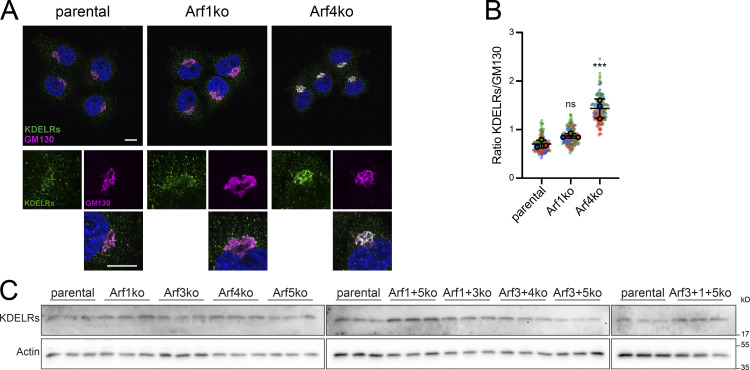
Functionality of KDEL-mediated retrieval in Arf KO cell lines was tested by transient expression of a signal sequence–GFP–KDEL (GFP-KDEL) construct and examination of its steady-state localization (Fig. 5 D). In parental HeLaα cells, the GFP signal was visible in the reticular pattern of the ER and a number of puncta. Exclusively in cells lacking Arf4, the GFP-KDEL–positive puncta were completely lost and the reticular staining reduced. Instead, a perinuclear accumulation colocalizing with GM130 was detected, consistent with a defective retrieval from the Golgi back to the ER in the absence of Arf4.
Therefore, we determined the localization of the KDEL receptors in Arf1ko and Arf4ko cells, which showed the strongest defects in βCOP recruitment and GFP-KDEL transport, respectively (Fig. S5 A). In parental HeLaα and Arf1ko cells, the KDEL receptor staining exhibited a faint perinuclear staining colocalizing with the GM130 signal and puncta distributed throughout the cell. In Arf4ko cells, the signal appeared more intense in the perinuclear region. Quantitation of Golgi-localized KDEL receptor signal relative to that of GM130 (Fig. S5 B) confirmed a significant increase of KDEL receptors at the Golgi in Arf4ko cells compared with parental cells. This change was caused by altered transport, since the intracellular levels of KDEL receptors remained unchanged upon Arf4 KO (Fig. S5 C).
Figure S5.
Intracellular localization and protein level of KDEL receptors. (A) Parental HeLaα, Arf1ko, and Arf4ko cells were coimmunostained for KDEL receptors (KDELRs) in green and GM130 in magenta. DAPI signal is shown in blue. Lower panels show magnified image sections. Scale bars, 10 µm. (B) Perinuclear localization of the KDEL receptors was quantified as a ratio to GM130. Average values obtained from three independent experiments with >40 Golgi analyzed per cell line and experiment are shown as filled circles together with the values from individual cells as small dots colored by experiment. Mean ± SD of the three experiments are indicated. Unpaired one-way ANOVA versus parental (***, P < 0.001). (C) KDEL receptor levels in parental HeLaα and Arf KO cell lines were analyzed by immunoblot analysis with actin as a loading control. For each cell line, three biological replicates were analyzed on the same gels with parental lysates.
Defective retrieval and aberrant secretion of proteins with a KDEL motif and accumulation of KDEL receptors at the Golgi in cells lacking Arf4 indicate an important role for Arf4 in retrograde transport of KDEL receptors and their cargo from the Golgi to the ER.
Rescue experiments to define Arf specificity
To assess the specificity of the observed Arf1ko and Arf4ko phenotypes and the rescue potential of different Arfs, stable cell lines were generated by lentiviral transduction to overexpress individual Arfs in parental HeLaα, Arf1ko, and Arf4ko cells. Compared with the endogenous Arf levels in parental cells, Arf1 and Arf4 were moderately, and Arf3 and Arf5, which have low endogenous levels, were highly overexpressed (Fig. 6, A and B; and Fig. 7, A and B).
Figure 6.
Rescue experiments of the Arf1ko phenotype by overexpressing Arf1, Arf3, or Arf4. Based on parental HeLaα and Arf1ko cells, stable cell lines were generated by lentiviral transduction, either expressing the empty vector (+empty) as a control or overexpressing the indicated untagged Arf. (A and B) Expression levels of Arfs in clonal (A) and pooled cell lines (B) were analyzed by immunoblotting. Ten and five times less lysate were loaded for Arf-overexpressing cell lines in A and B, respectively. Actin served as a loading control. (C and D) Parental HeLaα and Arf1ko cell lines transduced with the empty vector or overexpressing class I Arfs Arf1 and Arf3 (C) or class II Arf Arf4 (D) were immunostained for GM130 to examine Golgi morphology. Magnified sections are shown in the lower panels. Scale bars, 10 µm.
Figure 7.
Rescue experiments of the Arf4ko phenotype by overexpressing Arf1, Arf4, or Arf5. Stable cell lines were generated from parental HeLaα and Arf4ko cells by lentiviral transduction, either expressing the empty vector (+empty) as a control or overexpressing the indicated untagged Arf. (A and B) Expression levels of Arfs in clonal (A) and pooled (B) cell lines were analyzed by immunoblotting. Ten and five times less lysate were loaded for Arf-overexpressing cell lines in A and B, respectively. In B, two irrelevant lanes were removed as indicated by the black line. Actin served as a loading control. (C and D) Immunoblot analysis probing the media (M) and cell lysates (L) for the chaperones BiP and calreticulin (Calret.) of parental HeLaα and Arf4ko cell lines transduced with the empty vector or overexpressing class II Arfs (C) or class I Arf Arf1 (D). Actin served as a loading control. (E and F) Immunofluorescent microscopy revealed the steady-state localization of transiently expressed signal sequence–GFP–KDEL (green) coimmunostained with GM130 (magenta) in parental HeLaα and Arf4ko cell lines transduced with the empty vector or overexpressing class II Arfs Arf4 or Arf5 (E) or overexpressing class I Arf Arf1 (F). DAPI signal is shown in blue. Lower panels show magnified image sections. Asterisks mark images with enhanced intensity. Scale bars, 10 µm.
Rescue of the Arf1ko phenotype was assessed based on Golgi morphology visualized by immunofluorescence staining of GM130. In parental HeLaα cells, overexpression of Arf1 or the other class I member, Arf3, showed an opposite effect of Arf1 deletion: the Golgi condensed onto individual filaments and puncta (Fig. 6 C). The same phenotype was also obtained in Arf1ko cells, indicating reversion of the KO phenotype. Overexpression of Arf4, the main class II Arf, in Arf1ko cells also resulted in a condensation of the Golgi (Fig. 6 D).
The measure for rescue of the Arf4ko phenotype was the restoration of ER protein retrieval. As demonstrated by immunoblot analysis, aberrant secretion of BiP and calreticulin in Arf4ko cells was completely reverted to background levels upon reexpression of Arf4 and also partially upon overexpression of Arf5, the other class II Arf (Fig. 7 C). No effect of Arf4 overexpression was observed in control HeLaα cells. Interestingly, Arf5 overexpression in parental HeLaα cells caused significant secretion of these ER chaperones. We speculate that Arf5 cannot completely substitute for Arf4 with respect to ER protein retrieval, and if Arf4 is present, overexpressed Arf5 competes with Arf4, but is less productive. Remarkably, overexpression of the main class I member Arf1 in Arf4ko cells did not ameliorate the retrieval of ER chaperones, and the level of secreted proteins remained comparable to Arf4ko cells (Fig. 7 D).
Immunofluorescence microscopy of GFP-KDEL confirmed these observations by full rescue of ER staining with punctate accumulations in Arf4ko cells reexpressing Arf4 and a slightly weaker rescue by Arf5 overexpression (Fig. 7 E). In Arf4ko cells overexpressing Arf1, the GFP-KDEL signal remained concentrated in the perinuclear region with no puncta visible (Fig. 7 F).
Reversal of the deletion phenotypes by reexpression confirms that they were caused by the particular KO. In both cases, overexpression of the other Arf of the same class produced similar results, although to different extents. This suggests at least partial redundancy between the Arfs of the same class. Overexpression of the main Arf of the other class in the KO cell lines indicated that the Arf1ko phenotype arises from a more general defect that can be compensated by other Arfs, while the Arf4ko phenotype is caused very specifically by the absence of Arf4.
Discussion
No single Arf is essential and Arf4 is sufficient for cell survival
Members of the Arf family are found in all known eukaryotes from yeast to human. Among the “true” Arfs, class I orthologues are present more frequently, while class II Arfs arose later in evolution (Gillingham and Munro, 2007; Donaldson and Jackson, 2011). In yeast, for instance, three Arfs exist, which are homologous to class I (Arf1 and Arf2) and class III (Arf3) Arfs, but none of class II (Lee et al., 1994). None of the three Arfs is essential, but the simultaneous deletion of both class I Arfs is lethal (Stearns et al., 1990).
Arf GTPases are important regulators of a range of cellular processes, especially within the secretory pathway. For this reason, their misregulation is associated with diverse human diseases (reviewed in Casalou et al., 2016; Sztul et al., 2019). In cancer, for instance, Arf signaling was reported to be altered, most commonly by gene amplification. Moreover, Arf4 and Arf6 were linked to cancer cell migration, invasion, and metastasis (Sztul et al., 2019). Hence, it is important to decipher and understand shared and specific functions of individual Arfs. Their identification has been complicated, for instance, by apparent functional redundancy, shared GEFs and GAPs, and a complex interactome, as well as technical difficulties (Sztul et al., 2019).
Previous cellular analyses employed shRNA- or siRNA-mediated knockdowns, potentially prone to incomplete depletion and off-target effects, or (over)expression of mutant Arfs. Dominant-active (GTPase-deficient) or dominant-negative (GDP-locked) mutants may interfere with the activity of other Arfs by outcompeting them or by blocking shared GEFs. Furthermore, epitope-tagging was shown to change the Arfs’ properties in subtle ways (Jian et al., 2010).
Our approach to gain insight into Arf functions was based on genomic deletions of individual Arfs and Arf combinations and subsequent observation of defects in cell growth, Golgi morphology, and coat recruitment to the Golgi. We successfully generated nine genomic Arf KO cell lines, composed of four single-KOs of each human class I (Arf1 and Arf3) and class II (Arf4 and Arf5) Arf, and four double-KO and one triple-KO combination. These cells are permanently depleted for the respective Arfs, which may lead to compensatory effects. In this line, we only observed an up-regulation of Arf4 in all cell lines lacking Arf1 (Fig. 1 B), but no further changes in Arf expression levels.
In agreement with knockdown based studies, we found that cultured cells can cope rather well with the loss of any single Arf and even certain combinations. However, the single-KO of Arf1 or Arf4 exhibited significant and distinct phenotypes, for example, either deletion reduced the growth rate and increased the fraction of cells in G0/1 phase of the cell cycle. The combined Arf1+4 double-KO was not viable. Interestingly, the same is true for simultaneous deletion of both class II Arfs (4+5), whereas cells knocked out for both class I Arfs (1+3) grew as well as Arf1ko cells. In addition, Arf4 was found to be able to sustain all essential Arf functions alone in cultured cells lacking the other three Arfs. Results may differ in other cell lines, as is documented by the loss-of-function screens in DepMap (http://www.depmap.org), which show that several cell lines depend on Arf1 and/or Arf4. Nontransformed cells may also be more sensitive to the lack of individual Arfs. In mice, KOs of Arf1 or Arf4 (as well as of Arf6) were embryonically lethal, and conditional KOs in specific tissues led to various dysfunctions (Sztul et al., 2019; International Mouse Phenotyping Consortium [http://www.mousephenotype.org]).
Deletion of Arf1 increases Golgi volume and reduces coat recruitment
The most striking phenotype of cells deleted for Arf1, alone or in combination with Arf3 and/or Arf5, was an enlargement of the Golgi, apparent in a more expanded GM130 staining, a higher volume in 3D Golgi reconstructions, and longer Golgi stack cross-sections in electron microscopy. Furthermore, steady-state densities of COPI, AP1, and GGA2 vesicle coat components at the Golgi were reduced. This indicates a lower rate of coat recruitment and thus of formation of COPI and AP1/GGA/clathrin transport vesicles per Golgi area, causing a reduction of cargo and membrane traffic originating from the Golgi in retrograde and anterograde direction. The resulting imbalance of influx and efflux might explain the increase in Golgi size, as proposed by Sengupta and Linstedt (2011). Indeed, the reduction in coat density was found to be compensated by the increase in Golgi size restoring the equilibrium between incoming and outgoing material.
An increase in Golgi volume has also been observed physiologically upon an increased demand for cargo transport, processing, and sorting at the Golgi (Sengupta and Linstedt, 2011). As cells grow during interphase, increased cargo load causes a Golgi volume increase. Golgi growth was shown in HeLa cells to occur by cisternal elongation of existing Golgi stacks rather than by addition of new stacks (Sin and Harrison, 2016). Thus, the observed length increase in Golgi stacks of our Arf1 KO cells is likely to result from increased cargo content due to reduced export rates. Taken together, Arf1 appears to be the major mediator of vesicle traffic originating from the Golgi. Similar observations were made in yeast, where KO of Arf1 also resulted in an increase in Golgi volume (Gaynor et al., 1998; Bhave et al., 2014; Iyer et al., 2018).
KO of Arf4 specifically disrupts retrieval of ER proteins
The deletion of Arf4, alone or in combination with Arf3, caused a slight increase in Golgi volume and a reproducible but not statistically significant decrease of coat components at the Golgi. Arf4 thus appears to contribute to the recruitment of COPI, AP1, and GGA2, and the consequential mild reduction of Golgi exit might explain the observed slight increase in Golgi volume in Arf4ko cell lines.
Most strikingly, however, Arf4ko cells manifest an aberrant secretion of ER resident proteins that are normally retrieved from the Golgi back to the ER by the KDEL receptors. The phenotype resembles the one reported for knockdowns of multiple KDEL receptors (Li et al., 2015). However, this defect upon Arf4 deletion cannot be simply linked to a general defect in retrograde transport due to reduced COPI recruitment, since this is more strongly observed in Arf1ko cells without causing ER protein secretion. Moreover, overexpression of Arf1, the main COPI-recruiting Arf, is unable to compensate for the loss of Arf4 in this function. Thus, a more specific function in ER protein retrieval must be defective in Arf4ko cells.
It has previously been shown that in addition to Arf1, the class II Arfs, but not Arf3, can competitively support COPI vesicle formation (Popoff et al., 2011). For COPI, it has been shown that two paralogs of γ-COP (γ1/γ2) and of ζ-COP (ζ1/ζ2) can form three distinct COPI complexes (γ1ζ1, γ1ζ2, or γ2ζ1) with potentially different functions (Moelleken et al., 2007; Wegmann et al., 2004). Furthermore, Scyl1, a member of the Scy1-like family of catalytically inactive protein kinases, was identified as an interactor of COPI at the cis-Golgi and ERGIC that causes reduced retrograde traffic of the KDEL receptors when inactivated (Burman et al., 2008, 2010). Subsequently, Scyl1 was shown to bind to class II Arfs, preferentially Arf4, and to interact directly with the COPI subunit γ2 (Hamlin et al., 2014). This interaction was recently shown to depend on arginine methylation of Scyl1 by PRMT1 (Amano et al., 2020). A tripartite Scyl1–Arf4–γ2ζ1-COP complex thus was proposed to specifically mediate KDEL receptor traffic. This mechanism may thus account for the specific phenotype we observe upon Arf4 deletion.
However, in other studies, Scyl1 was also reported to bind preferentially to Arf1 and to GORAB, a protein associated with gerodermia osteodysplastica, at the trans-Golgi to promote COPI recruitment (Witkos et al., 2019). Pathogenic GORAB mutations cause impairment of COPI-mediated retrieval of trans-Golgi enzymes and a deficit in glycosylation of secretory proteins. Based on their results, the authors suggest that there might be two separate pools of Scyl1, a GORAB-dependent one at the trans-Golgi and a pool at the cis-Golgi/ERGIC for distinct COPI functions.
The situation is further complicated by the recent finding that a mutation in γ1-COP (K652E), shown to cause defective humoral and cellular immunity, disrupted KDEL receptor binding to COPI, thus affecting KDEL receptor localization, increasing ER stress in activated T and B cells and apoptosis in activated T cells (Bainter et al., 2021). Thus, other, γ1-containing COPI complexes also appear to contribute to KDEL receptor sorting in these cells. How Arf4 specifically mediates KDEL receptor retrieval is therefore not entirely clear yet.
Arf KO combinations
In the majority of viable Arf double- or triple-KO cell lines, no additional phenotypes were detected beyond those of Arf1 or Arf4 single deletion regarding Golgi size and morphology, coat recruitment, or secretion of ER resident proteins. However, in Arf3+1ko cells, the loss of GGA2 from the Golgi was more pronounced than in Arf1ko cells. This suggests a functional overlap of Arf1 and Arf3 in the recruitment of GGA2, which is consistent with Arf3’s known preferential localization to the TGN and activation by the trans-Golgi GEF BIG1 (Manolea et al., 2010) and its ability to bind GGAs (Boman et al., 2000). The only exception is the additional observation of a dilated ER in the Arf3+1+5 KO cell line.
Previous knockdown-based studies reported phenotypes only upon simultaneous silencing of two Arfs (Volpicelli-Daley et al., 2005; Kondo et al., 2012; Nakai et al., 2013). KDEL-receptor localization, for example, was described to be enhanced at the Golgi upon double knockdown of Arf3+Arf4 (Volpicelli-Daley et al., 2005) and of Arf4+Arf5 (Volpicelli-Daley et al., 2005; Li et al., 2015). Our results attribute this phenotype solely to the deletion of Arf4. The same applies to a slight compaction of the Golgi observed upon knockdown of Arf4+Arf5 (Nakai et al., 2013).
In other cases, the reported phenotype, for instance the peripheral βCOP puncta observed upon knockdown of Arf1+Arf3 and Arf1+Arf5 (Volpicelli-Daley et al., 2005; Kondo et al., 2012), cannot be rationalized by our KOs. Knockdown of Arf combinations for which no KO cell lines could be generated could provide information on the defects that lead to growth arrest or cell death. In this line, the simultaneous knockdown of Arf1+Arf4 severely impacted Golgi morphology and AP1 and COPI localization (Volpicelli-Daley et al., 2005; Nakai et al., 2013).
In the present study, we established Arf KO cell lines as tools to study shared and specific functions of Arfs at the Golgi. Of course, these cell lines offer themselves to investigate Arf-dependent processes that do not require specialized cell types.
Materials and methods
Cell culture
Helaα cells were grown in DMEM (high glucose; Sigma-Aldrich) with 10% FCS (FCS premium, VWR), 2 mM L-glutamine, 100 U/ml penicillin G, and 100 ng/ml streptomycin at 37°C and 7.5% CO2.
HeLaα KO cell lines
Two gRNAs were designed for each targeted Arf gene as listed in Table S5 to facilitate genomic deletion of exon 1 using several online tools (e.g., CRISPOR; Concordet and Haeussler, 2018). gRNAs were cloned in the pSpCas9(BB)-2A-GFP (Addgene plasmid #48138; a gift from Feng Zhang, Broad Institute, Cambridge, MA), and pSpCas9(BB)-2A-mCherry, which was derived the former by exchanging GFP to mCherry. Target cells were transfected simultaneously with the corresponding plasmids using jetPRIME (Polyplus Transfection). After 24 h, double-fluorescent cells were selected by FACS (FACS AriaIII; BD Biosciences). After 7 d, double-negative cells were selected by FACS and single cells sorted into 96-well plates with growth medium containing 10% conditioned medium. After ∼14 d, clonal cell lines were expanded and analyzed.
Genomic PCR
Trypsinized cells were pelleted, resuspended in 10 mM Tris (pH 8.7), heated at 95°C for 10 min, incubated with proteinase K (0.5 µg/µl) for 20 min at 37°C, inactivated at 95°C for 15 min, and used as DNA template for PCR with Phusion Polymerase (NEB) or Q5 Polymerase (NEB, for Arf5 KOs), following the manufacturer´s protocol for high GC content. Primers designed to amplify the genomic region surrounding the site of deletion are listed in Table S5.
Immunoblot analysis
Cell lysates were denatured in SDS-sample buffer for 5 min at 95°C (10 min at 37°C to analyze KDEL receptors), separated by SDS-gel electrophoresis (15% polyacrylamide for Arfs), and transferred to Immobilon-P PDVF membranes (Millipore). Membranes were blocked with TBST (20 mM Tris, 150 mM NaCl, pH 7.6, and 0.1% Tween20) with 3% nonfat dry milk for 1 h and incubated with primary antibody in TBST with 1% milk overnight at 4°C: anti-Arf1 (1:2,500; MAB10011; Abnova), anti-Arf3 (1:1,000; 610784; BD Bioscience), anti-Arf4 (1:1,000; 11673–1-AP; Proteintech), anti-Arf5 (1:750; H00000381-M01; Abnova), anti-actin (1:100,000; MAB1501; Sigma-Aldrich), anti-calreticulin (1:2,500; 27298–1-AP; Proteintech), anti-Grp78/BiP (1:10,000; GTX113340-10; Genetex), and anti-KDEL receptor antibody (1:1,000; 69659; Abcam). After washing, the membranes were incubated with HRP-conjugated secondary antibody (1:10,000, anti-rabbit, A0545, Sigma-Aldrich; anti-mouse, A0168, Sigma-Aldrich) in TBST with 1% milk. Chemiluminescence signals were detected using Immobilon Western HRP Substrate (Millipore) or Radiance Plus (Azure Biosystems) and imaged using a FusionFX (Vilber Lourmat).
Growth assay
Cells were seeded in 12-well plates at a density of 5,500 cells/well, which was confirmed by recounting. Every 24 h for 5 consecutive days, cells from one well for each cell line were trypsinized, resuspended in PBS, and counted. Doubling times were estimated by exponential fitting of the growth curves.
Flow cytometry
The procedure was adapted from Sin and Harrison (2016). Trypsinized cells were collected in ice-cold 10% FCS in PBS, pelleted at 50 rcf (relative centrifugal force) for 5 min at 4°C, washed once with PBS, and resuspended in PBS. Cells were fixed with ice-cold 70% ethanol on ice for at least 2 h. Afterwards, cells were incubated in PBS for 15 min at room temperature and counted. Per condition, 1.5–2 · 106 cells were used. Cells were incubated in PBS with 200 µg/ml RNase A, 0.1% Triton X-100, and 2 µg/ml propidium iodide (Sigma-Aldrich). Fluorescence signals were measured by flow cytometry using the LSR Fortessa (BD Biosciences). Data were analyzed with the FlowJo software, first gating for single cells and subsequently applying the built-in cell cycle fitting model.
Immunofluorescence staining
Cells were grown on glass coverslips for 1 d, then fixed with 3% PFA for 10 min, quenched with 50 mM NH4Cl in PBS for 5 min, permeabilized with 0.2% Triton X-100 in PBS for 10 min, blocked with 1% BSA in PBS for 1 h, and incubated with primary antibodies diluted in 1% BSA in PBS for 1 h: anti-AP1γ1 (1:1,000; self-made from hybridoma cells), anti-βCOP (1:500; CM1; hybridoma supernatant; gift from Dr. Felix Wieland, Heidelberg University, Heidelberg, Germany), anti-GGA2 (1:500; 612613; BD Bioscience), anti-GM130 (1:1,000; 12480S; Cell Signaling), and anti-TGN46 (1:1,000; AHP500G; Bio-Rad). Samples were washed and incubated with fluorescent secondary antibodies diluted in 1% BSA in PBS for 1 h (1:400, anti-mouse-Alexa488, A21202, Invitrogen; anti-rabbit-Alexa568, A10042, Invitrogen; anti-sheep-Cy3, 713–165-147, Jackson ImmunoResearch). For KDEL receptor staining, cells were fixed and quenched as described above, permeabilized with 0.1% saponine in PBS for 20 min, and blocked in 2% BSA and 0.1% saponine in PBS for 30 min. KDEL receptor antibody (1:1,000; 69659; Abcam) and the secondary antibody were diluted in blocking solution and incubated for 1 h and 30 min, respectively. Coverslips were mounted in FluoromountG (SouthernBiotech) supplemented with 0.5 ng/ml DAPI (Sigma-Aldrich) for confocal microscopy and in Vectashield (Vector Laboratories) for super-resolution microscopy, and stored in the dark at 4°C. For localization of GFP-KDEL, cells were grown on coverslips for 1 d, transfected with pcDNA3-ss-GFP-KDEL, and fixed and stained a day later.
Confocal microscopy and quantitation of coat localization and Golgi volume
Images were acquired using a LSM700 Upright confocal laser-scanning microscope with the Zen 2010 software (Zeiss) equipped with a Plan-Apochromat 63×/1.4 oil-immersion objective lens and two photomultiplier tubes. Imaging parameters were kept constant throughout each experiment. For quantitation of coat proteins at the Golgi, the GM130-stained area was selected in Fiji using the freehand tool, and the mean fluorescence intensity was measured for GM130 and the coat protein. To measure Golgi volume, z-stacks at 0.13 µm per slice were acquired and analyzed in Fiji using the “3DGolgiCharacterization” script (https://doi.org/10.5281/zenodo.4068393).
Super-resolution microscopy
3D-SIM was performed on a DeltaVision OMX-Blaze V4 system (Cytiva) equipped with solid-state lasers. Images were acquired using a Plan Apo N 60×, 1.42 NA oil immersion objective lens (Olympus), and four liquid-cooled sCMOS cameras (pco.edge 5.5, full frame 2,560 × 2,160; PCO). Exciting light was directed through a movable optical grating to generate a fine-striped interference pattern on the sample plane. The pattern was shifted laterally through five phases and three angular rotations of 60° for each z-section. The 405-, 488-, and 568-nm laser lines were used during acquisition, and the optical z-sections were separated by 0.125 µm. For the acquisition at 405 nm, laser power was attenuated to 50% with an exposure time of 40 ms, for 488 nm to 10% and 6 ms, and for 568 nm to 10% and 50 ms. Settings were adjusted to achieve optimal intensities of between 5,000 and 8,000 counts in a raw image of 15-bit dynamic range at the lowest laser power possible to minimize photobleaching. Multichannel imaging was achieved through sequential acquisition of wavelengths by separate cameras.
Raw 3D-SIM images were processed and reconstructed using the DeltaVision OMX SoftWoRx software package (v6.1.3; Cytiva). The resulting size of the reconstructed images was of 512 × 512 pixels from an initial set of 256 × 256 raw images. The channels were aligned in the image plane and around the optical axis using predetermined shifts as measured using a target lens and the SoftWoRx alignment tool. The channels were then carefully aligned using the alignment parameter from control measurements with 0.5-µm-diameter multi-spectral fluorescent beads (Invitrogen, Thermo Fisher Scientific). For visualization of the Golgi surface, we used the surface tool of the Imaris Cell Imaging software (Oxford Instruments).
Electron microscopy
Cells were fixed in serum-free medium with 2.5% glutaraldehyde and 3% formaldehyde for 2 h at room temperature, washed with PBS, and incubated with 2% osmium tetroxide and 1% K-hexacyanoferrate in H2O for 1 h at 4°C. After washing with H2O, uranyl-acetate (2% in H2O) was added and incubated at 4°C overnight. Cells were scraped after washing with H2O, pelleted, dehydrated by sequential incubation in 20%, 50%, 70%, and 90%, and three times 100% acetone/H2O for 30 min each, infiltrated with EMbed-812 (Electron Microscopy Science) according to the manufacturer’s protocol, and allowed to polymerize for 24 h at 60°C. The embedded cell pellets were cut into 60–70-nm-thin sections using an ultramicrotome (UltracutE, Reichert-Jung), collected on carbon-coated Formvar-Ni grids (Electron Microscopy Science), and stained for 10 min in 4% uranyl acetate and 2 min with lead citrate. Images were acquired on a CM100 electron microscope (Philips).
Analysis of secreted proteins
Cells were washed with PBS and incubated for 1 h with serum-free medium in the incubator. After another wash, secreted proteins were collected in a small volume of serum-free medium for 1 h. The collected medium was cleared by centrifugation at 10,000 rcf for 3 min at 4°C. Proteins were precipitated by the addition of 0.25 vol TCA, pelleted at 20,000 rcf at 4°C for 15 min, and washed with ice-cold acetone twice. For SDS-gel electrophoresis, these pellets and post-nuclear supernatants of cell lysates were boiled in SDS-gel sample buffer at 95°C for 10 min. Gels were stained with Commassie-R or analyzed by immunoblotting.
For mass spectrometry, 50 µl 2 M guanidium hydrochloride, 0.2 M Hepes, pH 8.3, and 10 mM tris(2-carboxyethyl)phosphine was added, and the pellet was sonicated in a Bioruptor Pico cooled by Minichiller 300 (both Diagenode). Chloroacetamide was added to a final concentration of 15 mM, and samples were incubated for 10 min at 95°C with gentle agitation. The concentration of guanidium hydrochloride was reduced to 0.5 M by dilution with 100 mM ammonium bicarbonate, and the samples were digested with 0.5 µg sequencing-grade trypsin (Promega) overnight at 37°C. The peptides were purified using C18 columns (BioPureSPN Mini Proto 300 C18, Nest Group) according to the manufacturer’s instructions and dried.
Desalted peptides were resuspended in 0.1% aqueous formic acid and subjected to liquid chromatography with tandem mass spectrometry analysis using a Q Exactive Plus Mass Spectrometer coupled with an EASY-nLC 1000 (both Thermo Fisher Scientific) and a custom-made column heater set to 60°C. Peptides were resolved using a reverse-phased HPLC column (75 µm × 30 cm) packed in-house with C18 resin (ReproSil-Pur C18–AQ, 1.9 µm resin; Dr. Maisch GmbH) at a flow rate of 0.2 µl/min. The following stepwise gradient of buffers A (0.1% formic acid in water) and B (80% acetonitrile, 0.1% formic acid in water) was used for peptide separation: 5–10% buffer B over 5 min, 10–35% over 45 min, 35–50% over 10 min, and finally 50–95% B over 2 min, followed by 18 min at 95% B.
The mass spectrometer was operated in data dependent acquisition mode with a total cycle time of ∼1 s. Each MS1 scan was followed by high-collision dissociation of the 10 most abundant precursor ions with dynamic exclusion set to 45 s. For MS1, 3 · 106 ions were accumulated in the Orbitrap over a maximum time of 100 ms and scanned at a resolution of 70,000 full width at half maximum (at 200 m/z). MS2 scans were acquired at a target setting of 105 ions, maximum accumulation time of 100 ms, and a resolution of 35,000 full width at half maximum (at 200 m/z). Singly charged ions and ions with unassigned charge state were excluded from triggering MS2 events. The normalized collision energy was set to 27% and the mass isolation window to 1.4 m/z, and one microscan was acquired for each spectrum.
Acquired raw files were imported into Progenesis QI software (v2.0, Nonlinear Dynamics Limited) to extract peptide precursor ion intensities across all samples applying the default parameters. The generated mgf file was searched using MASCOT against a human database (consisting of 41,484 forward and reverse protein sequences downloaded from UniProt on 20200417) and 392 commonly observed contaminants using the following search criteria: full tryptic specificity was required (cleavage after lysine or arginine residues, unless followed by proline); three missed cleavages were allowed; carbamidomethylation was set as fixed modification; oxidation and protein N-terminal acetylation were applied as variable modifications; mass tolerance of 10 ppm (precursor) and 0.02 D (fragments). The database search results were filtered using the ion score to set the false discovery rate to 1% based on the number of reverse protein sequence hits in the dataset. Results from label-free quantitation were processed using the SafeQuant R package v.2.3.2 (PMID: 27345528) to obtain peptide relative abundances. This analysis included global data normalization by equalizing the total peak/reporter areas across all liquid chromatography with mass spectrometry runs, data imputation using the knn algorithm, summation of peak areas per protein and liquid chromatography with tandem mass spectrometry run, followed by calculation of peptide abundance ratios. Only isoform specific peptide ion signals were considered for quantification. To meet additional assumptions (normality and homoscedasticity) underlying the use of linear regression models and t tests, mass spectrometry–intensity signals were transformed from the linear to the log-scale. The summarized peptide expression values were used for statistical testing of between condition differentially abundant peptides. Here, empirical Bayes-moderated t tests were applied, as implemented in the R/Bioconductor limma package (PMID: 25605792). The resulting per protein and condition comparison P values were adjusted for multiple testing using the Benjamini–Hochberg method. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD028846 (https://www.ebi.ac.uk/pride/archive/projects/PXD028846). GOterm enrichment analysis was performed using GORILLA (Eden et al., 2007; 2009).
Lentiviral transduction
RNA was isolated from HeLaα cells using the RNeasy Mini Kit (Qiagen) and the RNase-Free DNase Set (Qiagen), and cDNA was reverse-transcribed using the IScript cDNA Synthesis Kit (Bio-Rad). The coding sequences of Arfs were amplified by PCR (primers listed in Table S5), inserted into the pQXCIP plasmid (Takara Bio) using the AgeI and BamHI restriction sites, and sequenced. Plasmids were transfected into the packaging cell line Phoenix-ampho (Nolan laboratory, Stanford University, Stanford, CA) using jetPRIME (Polyplus Transfection). After 24 h, medium was exchanged to a smaller volume of medium supplemented with 1 mM pyruvate. Medium containing the viral particles was collected after 36 h and cleared by filtration through a 0.45-µm filter. After addition of polybrene to 20 µg/ml, the supernatant was transferred onto target cells. Selection was started 48 h after transduction using medium containing 1.5 mg/ml puromycin (InvivoGen). After 10 d, single cells were sorted by FACS (FACS AriaIII) into 96-well plates with puromycin-containing growth medium supplemented with 10% conditioned medium. Clonal cell lines with comparable expression levels in parental and KO background were selected based on immunoblotting for analysis (Fig. 6, A and C; and Fig. 7, A, C, and E). Alternatively, pooled cells obtained after 10 d of puromycin selection were used (Fig. 6, B and D; and Fig. 7, B, D, and F).
Statistics
SuperPlots were generated according to Lord et al. (2020), and statistical analysis was done with Prism8 (GraphPad) using unpaired or paired one-way ANOVA and unpaired, two-tailed t tests, respectively. Data distribution was assumed to be normal, but this was not formally tested.
Online supplemental material
Fig. S1 shows the cell cycle distribution of HeLaα, Arf1ko, and Arf4ko cells. Fig. S2 shows Golgi polarity in parental and Arf1ko cells by super-resolution microscopy. Fig. S3 displays the levels of βCOP in Arf KO cell lines by immunoblotting. Fig. S4 shows immunofluorescence colocalization of AP1γ1 and GGA2 with TGN46. Fig. S5 documents the localization and levels of the KDEL receptors in Arf KO cells. Table S1 lists gRNAs and primers. Table S2 lists significantly up-regulated hits of Arf4ko versus HeLa. Table S3 lists significantly up-regulated hits of Arf3+4 versus HeLaα. Table S4 lists GOterm enrichment analysis of Arf4ko versus HeLaα. Table S5 lists GOterm enrichment analysis of Arf3+4ko versus HeLaα.
Supplementary Material
lists gRNAs and primers.
lists significantly up-regulated hits of Arf4ko versus HeLa.
lists significantly up-regulated hits of Arf3+4 versus HeLaα.
lists GOterm enrichment analysis of Arf4ko versus HeLaα.
lists GOterm enrichment analysis of Arf3+4ko versus HeLaα.
Acknowledgments
We thank the Imaging Core Facility (University of Basel), in particular Dr. Kai Schleicher, Dr. Alexia Ferrand, and Laurent Guerard, for assistance at the microscopes and with data analysis, and Janine Bögli and Stella Stefanova of the FACS Core Facility for their support.
This work was supported by the Swiss National Science Foundation (grant 31003A-182519).
The authors declare no competing financial interests.
Author contributions: M. Pennauer: conceptualization, investigation, validation, visualization, writing – original draft/review, and editing. K. Buczak: investigation (mass spectrometry analysis). C. Prescianotto-Baschong: investigation (electron microscopy). M. Spiess: funding acquisition, conceptualization, project administration, resources, supervision, writing – original draft/review, and editing.
References
- Amano, G., Matsuzaki S., Mori Y., Miyoshi K., Han S., Shikada S., Takamura H., Yoshimura T., and Katayama T.. 2020. SCYL1 arginine methylation by PRMT1 is essential for neurite outgrowth via Golgi morphogenesis. Mol. Biol. Cell. 31:1963–1973. 10.1091/mbc.E20-02-0100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonny, B., Beraud-Dufour S., Chardin P., and Chabre M.. 1997. N-terminal hydrophobic residues of the G-protein ADP-ribosylation factor-1 insert into membrane phospholipids upon GDP to GTP exchange. Biochemistry. 36:4675–4684. 10.1021/bi962252b [DOI] [PubMed] [Google Scholar]
- Austin, C., Boehm M., and Tooze S.A.. 2002. Site-specific cross-linking reveals a differential direct interaction of class 1, 2, and 3 ADP-ribosylation factors with adaptor protein complexes 1 and 3. Biochemistry. 41:4669–4677. 10.1021/bi016064j [DOI] [PubMed] [Google Scholar]
- Bainter, W., Platt C.D., Park S.-Y., Stafstrom K., Wallace J.G., Peters Z.T., Massaad M.J., Becuwe M., Salinas S.A., Jones J., et al. 2021. Combined immunodeficiency due to a mutation in the γ1 subunit of the coat protein I complex. J. Clin. Invest. 131:140494. 10.1172/JCI140494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baschieri, F., and Farhan H.. 2012. Crosstalk of small GTPases at the Golgi apparatus. Small GTPases. 3:80–90. 10.4161/sgtp.19842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhave, M., Papanikou E., Iyer P., Pandya K., Jain B.K., Ganguly A., Sharma C., Pawar K., Austin J. II, Day K.J., et al. 2014. Golgi enlargement in Arf-depleted yeast cells is due to altered dynamics of cisternal maturation. J. Cell Sci. 127:250–257. 10.1242/jcs.140996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boman, A.L., Zhang C., Zhu X., and Kahn R.A.. 2000. A family of ADP-ribosylation factor effectors that can alter membrane transport through the trans-Golgi. Mol. Biol. Cell. 11:1241–1255. 10.1091/mbc.11.4.1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burman, J.L., Bourbonniere L., Philie J., Stroh T., Dejgaard S.Y., Presley J.F., and McPherson P.S.. 2008. Scyl1, mutated in a recessive form of spinocerebellar neurodegeneration, regulates COPI-mediated retrograde traffic. J. Biol. Chem. 283:22774–22786. 10.1074/jbc.M801869200 [DOI] [PubMed] [Google Scholar]
- Burman, J.L., Hamlin J.N.R., and McPherson P.S.. 2010. Scyl1 regulates Golgi morphology. PLoS One. 5:e9537. 10.1371/journal.pone.0009537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casalou, C., Faustino A., and Barral D.C.. 2016. Arf proteins in cancer cell migration. Small GTPases. 7:270–282. 10.1080/21541248.2016.1228792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavenagh, M.M., Whitney J.A., Carroll K., Zhang C., Boman A.L., Rosenwald A.G., Mellman I., and Kahn R.A.. 1996. Intracellular distribution of Arf proteins in mammalian cells. Arf6 is uniquely localized to the plasma membrane. J. Biol. Chem. 271:21767–21774. 10.1074/jbc.271.36.21767 [DOI] [PubMed] [Google Scholar]
- Concordet, J.-P., and Haeussler M.. 2018. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 46(W1):W242–W245. 10.1093/nar/gky354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dascher, C., and Balch W.E.. 1994. Dominant inhibitory mutants of ARF1 block endoplasmic reticulum to Golgi transport and trigger disassembly of the Golgi apparatus. J. Biol. Chem. 269:1437–1448. 10.1016/S0021-9258(17)42277-0 [DOI] [PubMed] [Google Scholar]
- De Matteis, M.A., and Godi A.. 2004. PI-loting membrane traffic. Nat. Cell Biol. 6:487–492. 10.1038/ncb0604-487 [DOI] [PubMed] [Google Scholar]
- Donaldson, J.G., and Jackson C.L.. 2011. ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat. Rev. Mol. Cell Biol. 12:362–375. 10.1038/nrm3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden, E., Lipson D., Yogev S., and Yakhini Z.. 2007. Discovering motifs in ranked lists of DNA sequences. PLOS Comput. Biol. 3:e39. 10.1371/journal.pcbi.0030039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden, E., Navon R., Steinfeld I., Lipson D., and Yakhini Z.. 2009. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 10:48. 10.1186/1471-2105-10-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor, E.C., Chen C.Y., Emr S.D., and Graham T.R.. 1998. ARF is required for maintenance of yeast Golgi and endosome structure and function. Mol. Biol. Cell. 9:653–670. 10.1091/mbc.9.3.653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham, A.K., and Munro S.. 2007. The small G proteins of the Arf family and their regulators. Annu. Rev. Cell Dev. Biol. 23:579–611. 10.1146/annurev.cellbio.23.090506.123209 [DOI] [PubMed] [Google Scholar]
- Hamlin, J.N.R., Schroeder L.K., Fotouhi M., Dokainish H., Ioannou M.S., Girard M., Summerfeldt N., Melançon P., and McPherson P.S.. 2014. Scyl1 scaffolds class II Arfs to specific subcomplexes of coatomer through the γ-COP appendage domain. J. Cell Sci. 127:1454–1463. 10.1242/jcs.136481 [DOI] [PubMed] [Google Scholar]
- Hsu, V.W., Shah N., and Klausner R.D.. 1992. A brefeldin A-like phenotype is induced by the overexpression of a human ERD-2-like protein, ELP-1. Cell. 69:625–635. 10.1016/0092-8674(92)90226-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzhak, D.N., Tyanova S., Cox J., and Borner G.H.. 2016. Global, quantitative and dynamic mapping of protein subcellular localization. eLife. 5:e16950. 10.7554/eLife.16950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer, P., Bhave M., Jain B.K., RoyChowdhury S., and Bhattacharyya D.. 2018. Vps74p controls Golgi size in an Arf1-dependent manner. FEBS Lett. 592:3720–3735. 10.1002/1873-3468.13266 [DOI] [PubMed] [Google Scholar]
- Jackson, C.L., and Bouvet S.. 2014. Arfs at a glance. J. Cell Sci. 127:4103–4109. 10.1242/jcs.144899 [DOI] [PubMed] [Google Scholar]
- Jian, X., Cavenagh M., Gruschus J.M., Randazzo P.A., and Kahn R.A.. 2010. Modifications to the C-terminus of Arf1 alter cell functions and protein interactions. Traffic. 11:732–742. 10.1111/j.1600-0854.2010.01054.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn, R.A., and Gilman A.G.. 1984. Purification of a protein cofactor required for ADP-ribosylation of the stimulatory regulatory component of adenylate cyclase by cholera toxin. J. Biol. Chem. 259:6228–6234. 10.1016/S0021-9258(20)82130-9 [DOI] [PubMed] [Google Scholar]
- Kahn, R.A., and Gilman A.G.. 1986. The protein cofactor necessary for ADP-ribosylation of Gs by cholera toxin is itself a GTP binding protein. J. Biol. Chem. 261:7906–7911. 10.1016/S0021-9258(19)57489-0 [DOI] [PubMed] [Google Scholar]
- Kahn, R.A., Cherfils J., Elias M., Lovering R.C., Munro S., and Schurmann A.. 2006. Nomenclature for the human Arf family of GTP-binding proteins: ARF, ARL, and SAR proteins. J. Cell Biol. 172(5):645–650. 10.1083/jcb.200512057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo, Y., Hanai A., Nakai W., Katoh Y., Nakayama K., and Shin H.-W.. 2012. ARF1 and ARF3 are required for the integrity of recycling endosomes and the recycling pathway. Cell Struct. Funct. 37:141–154. 10.1247/csf.12015 [DOI] [PubMed] [Google Scholar]
- Lee, F.J., Stevens L.A., Kao Y.L., Moss J., and Vaughan M.. 1994. Characterization of a glucose-repressible ADP-ribosylation factor 3 (ARF3) from Saccharomyces cerevisiae. J. Biol. Chem. 269:20931–20937. 10.1016/S0021-9258(17)31911-7 [DOI] [PubMed] [Google Scholar]
- Lewis, M.J., and Pelham H.R.. 1990. A human homologue of the yeast HDEL receptor. Nature. 348:162–163. 10.1038/348162a0 [DOI] [PubMed] [Google Scholar]
- Lewis, M.J., and Pelham H.R.. 1992. Sequence of a second human KDEL receptor. J. Mol. Biol. 226:913–916. 10.1016/0022-2836(92)91039-R [DOI] [PubMed] [Google Scholar]
- Li, M.Y., Grandadam M., Kwok K., Lagache T., Siu Y.L., Zhang J.S., Sayteng K., Kudelko M., Qin C.F., Olivo-Marin J.-C., et al. 2015. KDEL Receptors Assist Dengue Virus Exit from the Endoplasmic Reticulum. Cell Rep. 10:1496–1507. 10.1016/j.celrep.2015.02.021 [DOI] [PubMed] [Google Scholar]
- Li, Y., Kelly W.G., J.M. Logsdon, Jr., Schurko A.M., Harfe B.D., Hill-Harfe K.L., and Kahn R.A.. 2004. Functional genomic analysis of the ADP-ribosylation factor family of GTPases: phylogeny among diverse eukaryotes and function in C. elegans. FASEB J. 18(15):1834–1850. 10.1096/fj.04-2273com [DOI] [PubMed] [Google Scholar]
- Liang, J.O., and Kornfeld S.. 1997. Comparative activity of ADP-ribosylation factor family members in the early steps of coated vesicle formation on rat liver Golgi membranes. J. Biol. Chem. 272:4141–4148. 10.1074/jbc.272.7.4141 [DOI] [PubMed] [Google Scholar]
- Lord, S.J., Velle K.B., Mullins R.D., and Fritz-Laylin L.K.. 2020. SuperPlots: Communicating reproducibility and variability in cell biology. J. Cell Biol. 219:e202001064. 10.1083/jcb.202001064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolea, F., Chun J., Chen D.W., Clarke I., Summerfeldt N., Dacks J.B., and Melançon P.. 2010. Arf3 is activated uniquely at the trans-Golgi network by brefeldin A-inhibited guanine nucleotide exchange factors. Mol. Biol. Cell. 21:1836–1849. 10.1091/mbc.e10-01-0016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno-Yamasaki, E., Rivera-Molina F., and Novick P.. 2012. GTPase networks in membrane traffic. Annu. Rev. Biochem. 81:637–659. 10.1146/annurev-biochem-052810-093700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moelleken, J., Malsam J., Betts M.J., Movafeghi A., Reckmann I., Meissner I., Hellwig A., Russell R.B., Söllner T., Brügger B., and Wieland F.T.. 2007. Differential localization of coatomer complex isoforms within the Golgi apparatus. Proc. Natl. Acad. Sci. USA. 104:4425–4430. 10.1073/pnas.0611360104 [DOI] [PubMed] [Google Scholar]
- Nakai, W., Kondo Y., Saitoh A., Naito T., Nakayama K., and Shin H.W.. 2013. ARF1 and ARF4 regulate recycling endosomal morphology and retrograde transport from endosomes to the Golgi apparatus. Mol. Biol. Cell. 24:2570–2581. 10.1091/mbc.e13-04-0197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrotek, A., Zeghouf M., and Cherfils J.. 2016. Allosteric regulation of Arf GTPases and their GEFs at the membrane interface. Small GTPases. 7:283–296. 10.1080/21541248.2016.1215778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer, D.J., Helms J.B., Beckers C.J., Orci L., and Rothman J.E.. 1993. Binding of coatomer to Golgi membranes requires ADP-ribosylation factor. J. Biol. Chem. 268:12083–12089. 10.1016/S0021-9258(19)50311-8 [DOI] [PubMed] [Google Scholar]
- Popoff, V., Langer J.D., Reckmann I., Hellwig A., Kahn R.A., Brügger B., and Wieland F.T.. 2011. Several ADP-ribosylation factor (Arf) isoforms support COPI vesicle formation. J. Biol. Chem. 286:35634–35642. 10.1074/jbc.M111.261800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raykhel, I., Alanen H., Salo K., Jurvansuu J., Nguyen V.D., Latva-Ranta M., and Ruddock L.. 2007. A molecular specificity code for the three mammalian KDEL receptors. J. Cell Biol. 179:1193–1204. 10.1083/jcb.200705180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault, L., Guibert B., and Cherfils J.. 2003. Structural snapshots of the mechanism and inhibition of a guanine nucleotide exchange factor. Nature. 426:525–530. 10.1038/nature02197 [DOI] [PubMed] [Google Scholar]
- Sengupta, D., and Linstedt A.D.. 2011. Control of organelle size: the Golgi complex. Annu. Rev. Cell Dev. Biol. 27:57–77. 10.1146/annurev-cellbio-100109-104003 [DOI] [PubMed] [Google Scholar]
- Serafini, T., Orci L., Amherdt M., Brunner M., Kahn R.A., and Rothman J.E.. 1991. ADP-ribosylation factor is a subunit of the coat of Golgi-derived COP-coated vesicles: a novel role for a GTP-binding protein. Cell. 67:239–253. 10.1016/0092-8674(91)90176-Y [DOI] [PubMed] [Google Scholar]
- Sin, A.T.W., and Harrison R.E.. 2016. Growth of the Mammalian Golgi Apparatus during Interphase. Mol. Cell. Biol. 36:2344–2359. 10.1128/MCB.00046-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamnes, M.A., and Rothman J.E.. 1993. The binding of AP-1 clathrin adaptor particles to Golgi membranes requires ADP-ribosylation factor, a small GTP-binding protein. Cell. 73:999–1005. 10.1016/0092-8674(93)90277-W [DOI] [PubMed] [Google Scholar]
- Stearns, T., Willingham M.C., Botstein D., and Kahn R.A.. 1990. ADP-ribosylation factor is functionally and physically associated with the Golgi complex. Proc. Natl. Acad. Sci. USA. 87:1238–1242. 10.1073/pnas.87.3.1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztul, E., Chen P.-W., Casanova J.E., Cherfils J., Dacks J.B., Lambright D.G., Lee F.S., Randazzo P.A., Santy L.C., Schürmann A., et al. 2019. ARF GTPases and their GEFs and GAPs: concepts and challenges. Mol. Biol. Cell. 30:1249–1271. 10.1091/mbc.E18-12-0820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsu, H., Yoshino K., Toda K., and Nakayama K.. 2002. GGA proteins associate with Golgi membranes through interaction between their GGAH domains and ADP-ribosylation factors. Biochem. J. 365:369–378. 10.1042/bj20020428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teal, S.B., Hsu V.W., Peters P.J., Klausner R.D., and Donaldson J.G.. 1994. An activating mutation in ARF1 stabilizes coatomer binding to Golgi membranes. J. Biol. Chem. 269:3135–3138. 10.1016/S0021-9258(17)41837-0 [DOI] [PubMed] [Google Scholar]
- Thomas, L.L., and Fromme J.C.. 2016. GTPase cross talk regulates TRAPPII activation of Rab11 homologues during vesicle biogenesis. J. Cell Biol. 215:499–513. 10.1083/jcb.201608123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub, L.M., Ostrom J.A., and Kornfeld S.. 1993. Biochemical dissection of AP-1 recruitment onto Golgi membranes. J. Cell Biol. 123:561–573. 10.1083/jcb.123.3.561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpicelli-Daley, L.A., Li Y., Zhang C.-J., and Kahn R.A.. 2005. Isoform-selective effects of the depletion of ADP-ribosylation factors 1-5 on membrane traffic. Mol. Biol. Cell. 16:4495–4508. 10.1091/mbc.e04-12-1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegmann, D., Hess P., Baier C., Wieland F.T., and Reinhard C.. 2004. Novel isotypic gamma/zeta subunits reveal three coatomer complexes in mammals. Mol. Cell. Biol. 24:1070–1080. 10.1128/MCB.24.3.1070-1080.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkos, T.M., Chan W.L., Joensuu M., Rhiel M., Pallister E., Thomas-Oates J., Mould A.P., Mironov A.A., Biot C., Guerardel Y., et al. 2019. GORAB scaffolds COPI at the trans-Golgi for efficient enzyme recycling and correct protein glycosylation. Nat. Commun. 10:127. 10.1038/s41467-018-08044-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C.J., Rosenwald A.G., Willingham M.C., Skuntz S., Clark J., and Kahn R.A.. 1994. Expression of a dominant allele of human ARF1 inhibits membrane traffic in vivo. J. Cell Biol. 124:289–300. 10.1083/jcb.124.3.289 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists gRNAs and primers.
lists significantly up-regulated hits of Arf4ko versus HeLa.
lists significantly up-regulated hits of Arf3+4 versus HeLaα.
lists GOterm enrichment analysis of Arf4ko versus HeLaα.
lists GOterm enrichment analysis of Arf3+4ko versus HeLaα.