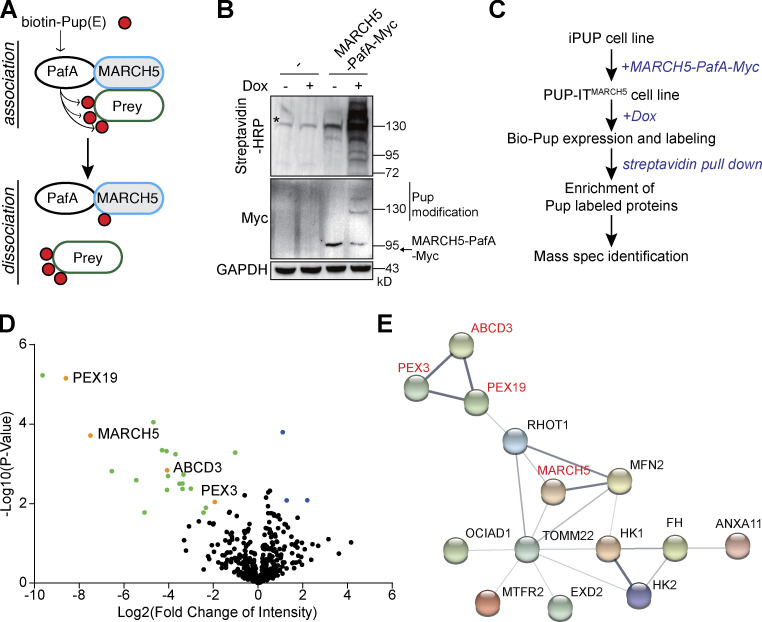
Figure 1.
Identification of MARCH5-interacting proteins by PUP-IT. (A) Design of PUP-ITMARCH5. Pup(E) ligase PafA was fused to the C-terminus of MARCH5 and catalyzed biotin-Pup(E) ligation to MARCH5-interacting proteins. (B) iPUP cells expressing MARCH5-PafA-Myc were treated with Dox for 24 h. iPUP cells without MARCH5-PafA-Myc were used as the control. Biotin-modified proteins were analyzed with streptavidin-HRP. The asterisk indicates background from endogenous biotin-modified proteins. (C) The workflow of PUP-ITMARCH5–based proximity labeling to identify MARCH5-interacting proteins. (D) Volcano plot of proteins enriched as MARCH5-interacting proteins. The logarithmic ratios of protein LFQ intensity (iPUP/PUP-ITMARCH5) were plotted against negative logarithmic P values from a two-sided, two-sample t test in Perseus. Green and orange dots represent proteins that are enriched in the PUP-ITMARCH5 sample (FDR ≤0.05; n = 3 independent experiments). (E) Proteins enriched in PUP-ITMARCH5 sample were subjected to gene ontology analysis. STRING (functional protein-association networks) was used to analyze protein interactions (https://string-db.org/). The thickness of the lines indicates the strength of data support. Spec, spectrometry.