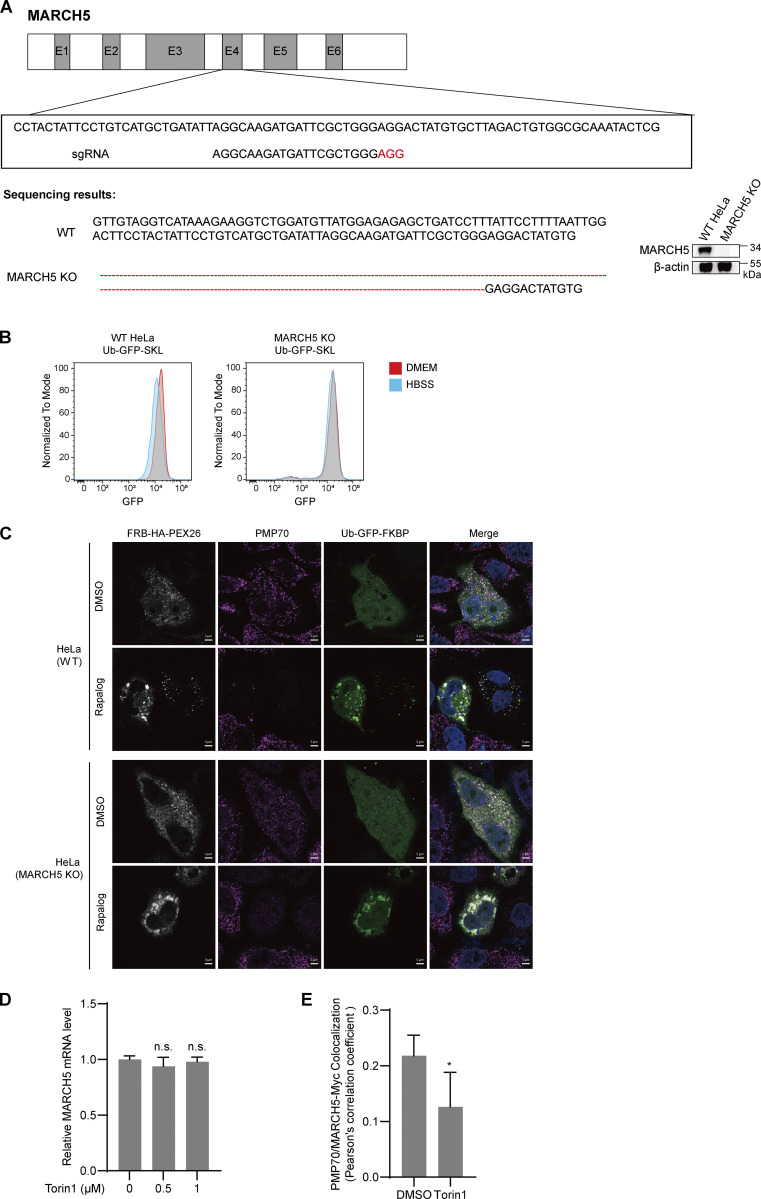
Figure S5.
Other supplementary information. (A) Schematic of the genome-editing strategy to knock out endogenous MARCH5 in HeLa cells. Exons 1–6 (E1-E6) are indicated. Three designed sgRNAs were tested initially, and the validated sgRNA confirmed by sequencing, as shown. Protospacer adjacent motif sequences are depicted in red. The KO of MARCH5 is confirmed by both Sanger sequencing (left) and IBs (right). (B) WT or MARCH5 KO HeLa cells expressing Ub-GFP-SKL were cultured in DMEM (supplemented with 10% FBS) or HBSS for 24 h. GFP signal was analyzed with FACS. (C) Representative images for Fig 9, D and E. WT or MARCH5 KO HeLa cells were transfected with Ub-GFP-FKBP and FRB-HA-PEX26 for 24 h. Cells were treated with DMSO or rapalog for 24 h, fixed, immunostained for PMP70, and imaged with confocal microscopy. Scale bars: 5 µm. (D) HeLa cells were treated with Torin1 (1 μM) for the indicated time. MARCH5 mRNA levels were detected with quantitative PCR assay. Values are mean ± SD, n = 3 independent experiments. n.s., not significant (P > 0.05) by two-tailed Student’s t test. (E) HeLa cells were transfected with MARCH5-Myc plasmid for 24 h, then with or without Torin1 (1 μM) were added for an additional 24 h before harvesting. Cells were then fixed and immunostained for PMP70 (Alexa Fluor 488, green) and Myc (Alexa Fluor 555, red) and examined by microscopy to detect GFP and RFP signals. Pearson’s correlation coefficients show the colocalization between PMP70 and MARCH5-Myc derived from above. Values are mean ± SD *, P < 0.05 by two-tailed Student’s t test.