Fig. 4.
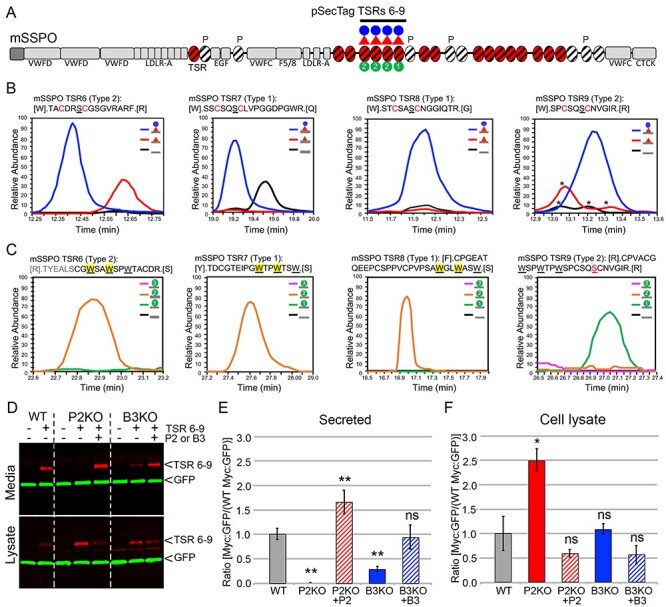
SSPO is a novel B3GLCT substrate and requires B3GLCT for efficient trafficking. (A) Domain structure of SCO-spondin (SSPO). The 25 SSPO TSRs are depicted by ovals. Red ovals identify 17 TSRs that have a consensus sequence for POFUT2 mediated O-fucosylation. White ovals are TSRs that lack a serine or threonine (S/T) in the consensus sequences for O-fucosylation, and P above a TSR indicates a proline in the position usually occupied by S/T. TSRs with consensus for modification with C-mannose are marked with diagonal black lines. In this study, we used the pSecTag2-SSPO TSRs 6–9 construct to confirm the presence of the glucose–fucose disaccharide (red triangle indicating fucose and blue circle glucose) and one or two C-mannose residues (green circles with #) on TSRs 6, 7, 8 and 9. Other SSPO domains include VWFD, von Willebrand factor–type D domain; LDLR-A, low density lipoprotein receptor class A domain; EGF, epidermal growth factor-like (EGF) domain; F5/8, factor V/VIII type C-like domain; VWFC, von Willebrand factor C repeats; CTCK, C-terminal cysteine knot domain. (B) Extracted ion chromatograms (EIC) of ions corresponding to the different glycoforms of peptides from TSRs 6, 7, 8 and 9 containing the POFUT2 consensus sequence. MS2 spectra for the major glycoforms and masses of ions used to generate the EICs can be found in Supplemental Figure 2. Although the score for the O-fucose glycopeptide from TSR6 was low, the mass of the parent ion matches the predicted mass of the glycopeptide and was used to generate the EIC. Black lines, unmodified peptide; red lines, fucose-modified peptide; blue lines, glucose–fucose–modified peptides. * indicate ions that match the mass of the relevant peptide but do not match the MS2 fragmentation pattern. (C) EICs of ions corresponding to the glycopeptides from TSRs 6, 7, 8 and 9 containing the C-mannosylation W-X-X-W motif. MS2 spectra for the major glycoforms and masses of ions used to generate the EICs can be found in Supplemental Figure 3. Black lines, unmodified peptide; green lines, Hex-modified peptide; orange lines, Hex–Hex modified peptides; pink lines, Hex–Hex–Hex modified peptides. The modified W is indicated by yellow shading for TSRs 6, 7 and 8. The MS2 data for the peptide from TSR9 are of insufficient quality to determine the location of the C-mannose. (D–F) Cell-based secretion assays were used to measure the effects of POFUT2 and B3GLCT mutations on trafficking of SSPO TSRs 6–9-Myc-His6 (TSRs 6–9, underlined in panel A) in wild-type HEK293T (WT), CRISPR-Cas9 mutagenized POFUT2 (P2 KO) or B3GLCT (B3 KO) HEK293T cells, or mutagenized cells rescued by cotransfection with plasmids encoding wild-type POFUT2 (+P2) or B3GLCT (+B3). (D) Representative western blot of the medium (above) and cell lysate (below). TSRs 6–9 were detected with anti-Myc (red), and GFP (green) was used as an internal transfection control. (E, F) Quantification of western blots of medium (E) and cell lysate (F). Data were evaluated for statistical significance using unpaired, one-tailed t-test. *P ≤ 0.05, **P ≤ 0.01, ns not significant. P ≤ 0.05).
