Fig. 3.
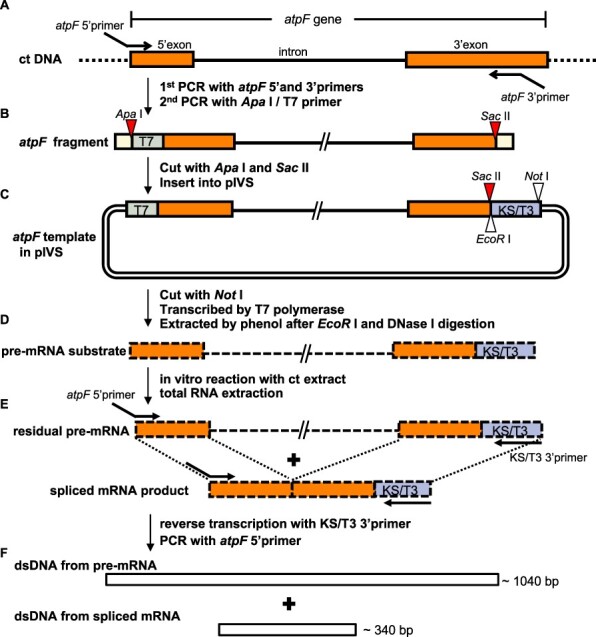
Schematic of the in vitro splicing procedures. (A) The atpF fragment was prepared from chloroplast (ct) DNA by a PCR amplification with the atpF 5′ primer (containing the 21-bp T7 promoter and the first 23 bp of the 5′ exon) and the atpF 3′ primer [containing a mid-3′ exon sequence (positions 100–137) and a SacII site]. The ApaI/T7 5′ primer was added and the PCR was continued to attach an ApaI site to the 5′ end. (B) The atpF fragment contains an ApaI site (red triangle), T7 (green), 145-bp full 5′ exon (orange), 695-bp intron (bold line), the first 137 bp of the 3′ exon (orange) and a SacII site (red triangle). Both ends (yellow) include either the full ApaI or SacII recognition sequence flanked by 4 nt (from the primers). The atpF fragment was digested with ApaI and SacII and then inserted into pIVS digested with the same enzymes. (C) The circular DNA (atpF.pIVS) was linearized by NotI and transcribed by T7 RNA polymerase. The resulting pre-mRNA was purified after digesting the template DNA with EcoRI and DNase I. (D) The pre-mRNA with a 37-bp KS/T3 sequence (blue) was incubated with a ct extract, after which the total RNA was extracted. (E) A RT was completed by adding the KS/T3 3′ primer (30 nt, complementary to KS/T3). A PCR amplification was then completed with the atpF 5′ primer. (F) The PCR products were separated by 2% agarose gel electrophoresis and stained with ethidium bromide. The expected sizes of the pre-mRNA (approximately 1,040 bp) and the spliced mRNA (approximately 340 bp) corresponded to the sequence from the end of the atpF 5′ primer to the end of the atpF 3′ primer. Primers are in Supplementary Fig. S8.
