ABSTRACT
Rapid and reliable detection of rifampin (RIF) resistance is critical for the diagnosis and treatment of drug-resistant and multidrug-resistant (MDR) tuberculosis. Discordant RIF phenotype/genotype susceptibility results remain a challenge due to the presence of rpoB mutations that do not confer high levels of RIF resistance, as have been exhibited in strains with mutations such as Ser450Leu. These strains, termed low-level RIF resistant, exhibit elevated RIF MICs compared to fully susceptible strains but remain phenotypically susceptible by mycobacterial growth indicator tube (MGIT) testing and have been associated with poor patient outcomes. Here, we assess RIF resistance prediction by whole-genome sequencing (WGS) among a set of 1,779 prospectively tested strains by both prevalence of rpoB gene mutation and phenotype as part of routine clinical testing during a 2.5-year period. During this time, 139 strains were found to have nonsynonymous rpoB mutations, 53 of which were associated with RIF resistance, including both low-level and high-level resistance. Resistance to RIF (1.0 μg/ml in MGIT) was identified in 43 (81.1%) isolates. The remaining 10 (18.9%) strains were susceptible by MGIT but were confirmed to be low-level RIF resistant by MIC testing. Full rpoB gene sequencing overcame the limitations of critical concentration phenotyping, probe-based genotyping, and partial gene sequencing methods. Universal clinical WGS with concurrent phenotypic testing provided a more complete understanding of the prevalence and type of rpoB mutations and their association with RIF resistance in New York.
KEYWORDS: Mycobacterium tuberculosis, WGS, rpoB, rpoA, rpoC, RIF, low-level resistance, MIC, rifampin, next-generation sequencing
INTRODUCTION
Rifampin (RIF) is show a critical component of drug regimens for treating tuberculosis (TB) infection, and detection of RIF resistance is crucial for the diagnosis and treatment of drug-resistant and multidrug-resistant (MDR) tuberculosis. RIF resistance is mainly caused by the presence of specific mutations in the rpoB gene, which encodes the RNA polymerase β subunit, an observation first described in Escherichia coli (1, 2). In 1993, Telenti et al. described the molecular basis for RIF resistance in 64 Mycobacterium tuberculosis clinical isolates and found resistance polymorphisms to be restricted to a highly conserved region of rpoB, a key finding that paved the way for the development of rapid genotypic methods for the determination of resistance (3, 4). This highly conserved region of rpoB would later be designated the 81-bp RIF resistance-determining region (RRDR) due to the frequency with which resistance polymorphisms were detected in this region (5, 6).
As genotypic methods to detect RIF resistance became widely implemented, rare variants associated with resistance were described, including several located outside the RRDR (7, 8). Among these less common variants, a class of mutations emerged that confer a lower level of RIF resistance than typical variants (9, 10). These mutations, referred to as low-level resistant mutations in this paper, were found to be susceptible to rifampin by critical concentration phenotypic susceptibility testing. It was proposed that these mutations are still clinically relevant (9, 11). This group of mutations has garnered increased attention since being linked to poor clinical outcomes when RIF is included in the treatment regimen (10, 12–15). Data regarding the prevalence of low-level RIF resistance in M. tuberculosis are still lacking, particularly in settings with low incidences of tuberculosis. Current reports suggest that the prevalence of these mutations vary by setting (12, 16, 17). Furthermore, mutations located outside the RRDR and rare variants within the RRDR can be challenging to detect or interpret, which may contribute to underreporting of these mutations, especially when some commonly used methods are used.
RIF resistance is most commonly found in conjunction with isoniazid (INH) resistance, but a minority of strains are resistant to only RIF and not INH or other TB drugs (RIF monoresistant). Reliable identification of RIF monoresistance is critical, as these strains have been associated with lower rates of successful treatment outcomes and may lead to the development of MDR-TB if treated suboptimally (18). The World Health Organization (WHO) estimated 1.1% of patients worldwide were infected with RIF monoresistant TB in 2014, but regional reports have suggested that the rate varies significantly, surpassing 20% in some jurisdictions (19, 20). RIF monoresistance has been reported at higher rates among HIV-positive patients compared to HIV-negative patients, while rates of resistance to other drugs, such as INH, have not been found to vary by HIV status (19, 21).
Studies have suggested that drug resistance in bacteria often results in a fitness cost, which may impact growth rate, virulence, and/or transmissibility (22, 23). In the case of rpoB mutations, compensatory mutations have been described in the rpoA and rpoC genes, which encode the α and β’ subunits of the RNA polymerase and have been suggested to compensate for the fitness cost of resistance mutations in rpoB (24). It has also been proposed that additional mutations in rpoB itself compensate for the fitness cost of resistance mutations, although these mutations are still not well characterized, likely due to the challenge discerning which mutations play a role in resistance versus compensation (23, 25). Reports of compensatory mutations are currently limited, but increased surveillance may further our understanding of how RIF resistance is acquired and spread.
To address these gaps in our knowledge, between January 2016 and October 2018 we tested one isolate of M. tuberculosis complex (MTBC) from each newly diagnosed patient in New York, a low-incidence setting (3.8 cases per 100,000) (26). Each isolate was tested by whole-genome sequencing (WGS), phenotypic susceptibility testing (Bactec mycobacterial growth indicator tube [MGIT] 960 and agar proportion), and selective MIC testing (Sensititre MYCOTB MIC plate). This report describes the incidence of low-level RIF mutations in M. tuberculosis strains in a prospective analysis in a low-incidence setting. Furthermore, we report on all rpoB gene mutations identified, their corresponding phenotypic interpretation, compensatory mutations, strain lineage, and genomic clustering among these strains.
MATERIALS AND METHODS
Clinical isolates.
A total of 1,779 MTBC strains from unique patients, received as isolates or cultured in-house from clinical specimens by the Mycobacteriology Laboratory at the Wadsworth Center, New York State Department of Health (NYSDOH), were included in this study. Prior to leaving the biosafety level 3 (BSL-3) laboratory for DNA extraction, liquid culture aliquots of clinical isolates were heat inactivated at 80°C for 60 min.
Phenotypic DST.
Culture-based RIF drug susceptibility testing (DST) was performed using the liquid culture MGIT 960 system (Bactec MGIT 960 SIRE package insert; Becton, Dickinson) and solid 7H10 agar proportion method according to the Clinical and Laboratory Standards Institute’s recommendations, using a critical concentration of 1.0 μg/ml (27). Isolates received in the laboratory were subcultured in MGIT medium prior to first-line DST being set up. A subset of 67 isolates was selected for MIC testing based on mutation profile and MGIT DST result. This group contained 53 isolates identified with rpoB gene mutations and a group of 14 wild-type strains. MIC testing was performed on cultures initially grown on Middlebrook 7H10 plates using the Sensititre MYCOTB AST plate (Thermo Scientific), including a RIF concentration range of 0.12 to 16 μg/ml. MIC testing was performed in triplicate, with plates being read by one analyst. Repeat MIC results were within 1 dilution for all isolates tested.
Real-time PCR.
An in-house-developed real-time PCR assay (28) was utilized to detect MTBC in all samples received. Prior to WGS, this assay was repeated as a quality control check on DNA extracts to assess purity and quantitation of MTBC DNA.
DNA extraction.
DNA was extracted from 1 ml of heat-inactivated isolates using a modified version of the InstaGene/FastPrep (IG/FP) method described by Shea et al. (29). Specifically, the 56°C incubation was reduced from 30 min to 10 min, and the volume of InstaGene matrix added to the pellets was altered to be 130 to 200 μl, based on the size of the pellet observed. These changes were implemented to reduce extraction turnaround time and increase DNA yield. DNA yields were measured by Qubit fluorometry and compared to real-time PCR results to confirm purity.
WGS.
Paired-end 250-bp DNA sequencing was carried out using the Illumina MiSeq platform following Nextera XT library preparation with a 15-cycle PCR indexing step (30). Sequencing runs were composed fully of 15 to 17 MTBC samples or of MTBC with other bacterial, viral, and/or parasitic samples.
Bioinformatic analysis.
Sequence analysis was performed using the Wadsworth Center TB WGS bioinformatics pipeline as previously described (29). Specifically, mutations were determined by a minimum 10× depth of coverage, and mutations were detected with the GATK package, using the diploid mode to allow for detection of heteroresistance. Major lineage identification was based on the presence or absence of lineage-defining single-nucleotide polymorphisms (SNPs; see Table S1 in the supplemental material), with nomenclature according to Gagneux and Small (31). Genomic clusters were determined by an SNP distance of ≤5 SNPs to any sequence(s) in our database, independent of epidemiological data.
Mutation classification.
Mutations in rpoB were sorted into three categories of RIF resistance, i.e., no resistance, low-level resistance, and high-level resistance, based on culture-based MGIT and MIC results. Strains with high-level resistance had clearly elevated RIF MIC (≥16 μg/ml) and were phenotypically resistant at 1.0 μg/ml in MGIT. Strains with low-level resistance had RIF MIC ranging from 0.25 to 1.0 μg/ml and tested phenotypically susceptible at 1.0 μg/ml in MGIT. Remaining mutations with no evidence of RIF resistance (susceptible at 1.0 μg/ml in MGIT and RIF MIC of ≤0.25 μg/ml) were determined not to be associated with RIF resistance. As the MIC ranges overlap at 0.25 μg/ml for low-level resistant and nonresistant strains, reports of mutations in the literature were used to finalize classification.
Data availability.
Sequences analyzed in this article have been provided to the Centers for Disease Control and Prevention on a monthly basis to contribute to the National TB Genotyping program (https://www.cdc.gov/tb/programs/genotyping/default.htm) as well as the Relational Sequencing TB Data Platform (ReSeqTB) (https://platform.reseqtb.org/), which catalogs a vast amount of genotypic, phenotypic, and related metadata from M. tuberculosis strains to enable the development of clinically useful, WHO-endorsed in vitro diagnostic assays for rapid drug susceptibility testing.
RESULTS
rpoB mutation identification and classification.
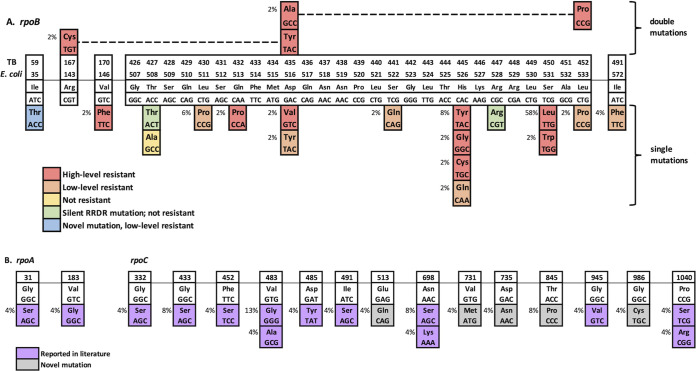
Among 1,779 isolates tested, 139 were found to have one or more nonsynonymous mutation(s) in the rpoB gene. A total of 119 had a single mutation, while the remaining 20 had two or three rpoB mutations each. Of the 139 strains, 53 (38%) were found to have mutations associated with phenotypic RIF resistance (both high- and low-level resistance) based on MGIT DST and MIC testing. These RIF-resistant isolates contained 17 specific mutations across 11 codons in rpoB (Fig. 1), including 42 (79.2%) MDR, 2 (3.8%) extensively drug-resistant (XDR), and 9 (17%) RIF monoresistant strains (Table 1). Nearly all RIF-resistant strains harbored mutations within the RRDR, 49/53 (92.5%), compared to 4/53 (7.5%) strains with single rpoB mutations outside the RRDR. Among the remaining 86 RIF-susceptible strains, four other mutations, including one nonsynonymous mutation (Thr427Ala) and three silent mutations (1 of Thr427Thr and 2 of Arg447Arg), were identified within the RRDR with no effect on resistance.
FIG 1.
Schematic representations of mutations in rpoB, rpoA, and rpoC genes. (A) rpoB. Cell color indicates corresponding phenotypic MGIT result. (B) rpoA and rpoC. Compensatory mutations among strains with rpoB gene resistance mutations. Cell color indicates previous literature reports. Lineage-specific mutations are not shown.
TABLE 1.
Compensatory mutations, susceptibility, and major lineage of strains with rpoB resistance mutationse
| Mutation | Description | No. resistant | No. sensitive | MIC | Resistance type | No. in major lineage: |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | ||||||||
| rpoB RRDR nonresistance SNPs | rpoB compensatory | rpoA | rpoC | ||||||||
| Thr427Thra | 0 | 1 | NA | Pan-susceptible | 1 | ||||||
| Thr427Ala | 0 | 1 | ≤0.12 | Pan-susceptible | 1 | ||||||
| Arg447Arga | 0 | 2 | NA | Pan-susceptible | 2 | ||||||
| rpoB low-level resistance mutations | rpoB compensatory | rpoA | rpoC | ||||||||
| Ile59Thrd | 0 | 1 | 0.5 | Mono | 1 | ||||||
| Leu430Pro | 0 | 3 | 0.5–1 | 1 MDR, 2 mono | 1 | 1 | 1 | ||||
| Asp435Tyr | 0 | 1 | 0.5 | MDR | 1 | ||||||
| Ser441Gln | 0 | 1 | NA | MDR | 1 | ||||||
| His445Gln | 0 | 1 | 0.25 | Mono | 1 | ||||||
| Leu452Pro | 0 | 1 | 1–2 | Mono | 1 | ||||||
| Ile491Phed | Thr845Pro | 0 | 2 | 1–2 | Mono | 2 | |||||
| rpoB high-level resistance mutations | rpoB compensatory | rpoA | rpoC | ||||||||
| Val170Phed | Val183Gly | 1 | 0 | N/Ab | MDR | 1 | |||||
| Gln432Pro | Gly31Ser | 1 | 0 | >16 | MDR | 1 | |||||
| Asp435Val | 1 | 0 | NAb | MDR | 1 | ||||||
| His445Cys | 1 | 0 | >16 | MDR | 1 | ||||||
| His445Gly | His674Arg | 1 | 0 | >16 | MDR | 1 | |||||
| His445Tyr | Val731Metc | 1 | 0 | >16 | MDR | 1 | |||||
| His445Tyr | Glu513Gln | 1 | 0 | MDR | 1 | ||||||
| His445Tyr | Glu563Ala | 2 | 0 | MDR | 2 | ||||||
| Ser450Trp | 1 | 0 | >16 | MDR | 1 | ||||||
| Ser450Leu | Gly332Ser | 1 | 0 | >16 | MDR | 1 | |||||
| Ser450Leu | Gly433Ser | 2 | 0 | MDR | 2 | ||||||
| Ser450Leu | Phe452Ser | 1 | 0 | MDR | 1 | ||||||
| Ser450Leu | Val483Ala | 1 | 0 | MDR | 1 | ||||||
| Ser450Leu | Val483Gly | 3 | 0 | MDR | 1 | 2 | |||||
| Ser450Leu | Asp485Tyr | 1 | 0 | MDR | 1 | ||||||
| Ser450Leu | Ile491Ser | 1 | 0 | MDR | 1 | ||||||
| Ser450Leu | Asn698Lys | 1 | 0 | MDR | 1 | ||||||
| Ser450Leu | Asn698Ser | 2 | 0 | MDR | 1 | 1 | |||||
| Ser450Leu | Asp735Asn | 1 | 0 | MDR | 1 | ||||||
| Ser450Leu | Gly945Val | 1 | 0 | MDR | 1 | ||||||
| Ser450Leu | Gly986Cys | 1 | 0 | MDR | 1 | ||||||
| Ser450Leu | Pro1040Ser | 1 | 0 | MDR | 1 | ||||||
| Ser450Leu | Pro1040Arg | 1 | 0 | MDR | 1 | ||||||
| Ser450Leu | 9 | 0 | 7 MDR, 2 Mono | 6 | 1 | 2 | |||||
| Ser450Leu | Arg552His | 1 | 0 | MDR | 1 | ||||||
| Ser450Leu | Asp574Glu | 1 | 0 | MDR | 1 | ||||||
| Ser450Leu | Val534Ala | 1 | 0 | XDR | 1 | ||||||
| Ser450Leu | Lys258Thr, Ser1039Ala |
1 | 0 | MDR | 1 | ||||||
| Double rpoB mutations | rpoB compensatory | rpoA | rpoC | ||||||||
| Arg167Cys, Asp435Tyr |
1 | 0 | >16 | XDR | 1 | ||||||
| Asp435Ala, Leu452Pro |
Asp265Gly | 1 | 0 | >16 | MDR | 1 | |||||
Silent mutation.
Sample exhausted; no MIC result.
Heterozygous mutation.
Mutation located outside RRDR.
TB numbering system used for rpoB gene. NA, not applicable.
MGIT DST, low-level resistance mutations, and RIF monoresistance.
Of the 53 strains with rpoB resistance mutations, 43 (81.9%) were resistant to RIF in MGIT at 1 μg/ml. The remaining 10 (18.9%) strains were susceptible in MGIT at 1 μg/ml but were determined to be low-level RIF resistant by MIC testing. Three of the mutations detected in these strains were located outside the RRDR (Ile59Thr [1], Ile491Phe [2]), and seven were within the RRDR (Leu430Pro [3], Asp435Tyr [1], Ser441Gln [1], His445Gln [1], Leu452Pro [1]). Two other strains were found to have mutations associated with low-level resistance, but each contained an additional resistance-associated mutation in rpoB, and both were phenotypically resistant to RIF in MGIT (Table 1). RIF monoresistance was more prevalent among low-level resistant strains (7/10, 70%) than high-level resistant strains (2/43, 4.6%). The 7 patients with low-level RIF monoresistant strains were HIV negative, while one of the two patients infected with high-level RIF monoresistant strains was HIV positive. HIV status for the remaining patient with high-level RIF monoresistant TB was unavailable.
MIC ranges.
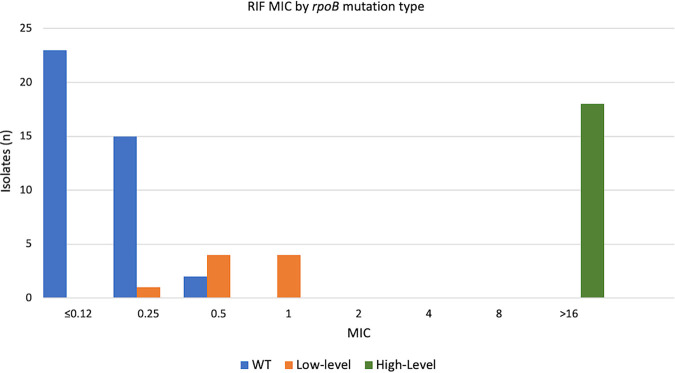
MIC results were performed for a total of 67 strains, including 53 with rpoB mutations and 14 wild-type strains. The results were grouped by rpoB mutation type: rpoB wild-type strains and rpoB mutations not associated with resistance were combined with a MIC range of <0.12 to 0.5 μg/ml (n = 40), rpoB mutation associated with low-level RIF resistance with a MIC range of 0.25 to 1.0 μg/ml (n = 9), and rpoB mutation associated with high-level RIF resistance with a MIC of >16 μg/ml (n = 18) (Fig. 2). Strains with high-level RIF resistance mutations had distinct MIC results compared to all other strains. The MIC ranges of rpoB wild-type and low-level rpoB mutated strains overlapped at 0.25 μg/ml and 0.5 μg/ml; one strain with rpoB His445Gln had a MIC of 0.25 μg/ml, while two rpoB wild-type strains had a MIC of 0.5 μg/ml.
FIG 2.
Rifampin MIC by rpoB gene mutation type: 67 strains, including 40 with no resistance mutations, 9 with low-level RIF resistance mutations, and 18 with high-level RIF resistance mutations.
Compensatory mutations.
Resistant strains were screened for the presence of potential compensatory mutations in the rpoA, rpoC, and rpoB genes. Thirty-two of 53 (60.4%) RIF-resistant strains harbored one of these mutations, 22 in rpoC, 8 in rpoB, and 2 in rpoA. Both rpoA mutations and 16/22 (73%) of the rpoC mutations were reported previously in RIF-resistant clinical isolates (Fig. 1). Mutations in compensatory genes were associated mainly with high-level RIF resistance; 30/43 (70%) of high-level RIF-resistant strains had a compensatory mutation compared to just 2/10 (20%) of low-level RIF-resistant strains. To our knowledge, we are the first to report rpoC mutations Glu513Gln, Val731Met, Asp735Asn, Thr845Pro, and Gly986Cys. We also considered all nonresistance SNPs in rpoB as potential compensatory mutations and found eight such mutations, not previously reported, in seven unique isolates with rpoB resistance mutations: Asp265Gly, Val534Ala, Arg552His, Glu563Ala, Asp574Glu, His674Arg, and Lys258Thr plus Ser1039Ala.
RIF resistance and mutation prevalence by major lineage.
All four major tuberculosis lineages (lineages 1 to 4) were represented in our collection (14.6%, 21.6%, 7.5%, and 51.7% of strains, respectively). Eighty-one strains were determined to be non-M. tuberculosis species in the MTBC (32 M. bovis BCG, 26 M. bovis, 18 M. africanum, 4 M. orygis, and 1 M. caprae), but none of these strains exhibited any RIF resistance. The distribution of strains with resistance-conferring rpoB mutations across the four major lineages of M. tuberculosis varied drastically, ranging from 5.7% in lineage 1 to 50.9% in lineage 2. In particular, resistance mutations in rpoB were overrepresented in lineage 2 strains, given the prevalence of these strains in our population (Table 2). Compensatory mutations in strains with rpoB resistance mutations were found at similar rates across each major lineage.
TABLE 2.
Major lineage and RIF resistance type summary
| Lineage | No. (%) of strains | No. (%) of strains with RIF resistancea | No. MDR/DR | No. RIF monoresistant | No. (%) of RIF-resistant strains with compensatory mutations |
|---|---|---|---|---|---|
| Lineage 1 (Indo-Oceanic) | 260 (14.6) | 3 (5.7) | 3 | 0 | 2 (67) |
| Lineage 2 (Beijing) | 384 (21.6) | 27 (50.9) | 23 | 4 | 16 (59) |
| Lineage 3 (Central Asian) | 134 (7.5) | 4 (7.6) | 4 | 0 | 3 (75) |
| Lineage 4 (Euro-American) | 920 (51.7) | 19 (35.8) | 14 | 5 | 11 (58) |
| Other lineagesb | 81 (4.6) | NA | NA | 0 | NA |
| Total | 1,779 | 53 | 44 | 9 | 32 |
Includes low-level and high-level RIF resistance.
Includes M. africanum, M. bovis, M. bovis BCG, M. orygis, and M. caprae.
Strain clustering.
In our study population, a SNP distance of ≤5 SNPs across the entire genome is suggestive of recent transmission based on follow-up epidemiological investigations (data not shown). WGS analysis identified three clusters among strains with rpoB resistance mutations. Cluster 1 was comprised of two strains with rpoB Ser450Leu and were genetically identical (0 SNPs). Clusters 2 and 3 included strains with rpoB Ile491Phe and His445Tyr, respectively. Each pair of strains in clusters 2 and 3 was separated by 3 SNPs. The remaining strains with rpoB resistance mutations did not belong to any genomic clusters. SNP-based estimates of recent transmission for strains with rpoB resistance mutations (5.7%) were lower than those for strains without rpoB resistance mutations (14.3%).
DISCUSSION
Since implementation in January 2016, routine, universal WGS in our laboratory has improved surveillance and detection of RIF resistance while providing insights into the background and characteristics of RIF-resistant strains in New York. In this study, we analyzed a prospective set of 1,779 MTBC isolates from unique patients over a 2.5-year period to determine the frequency and type of rpoB resistance mutations and their association with phenotypic resistance, including MIC testing. Additional data were collected to assess the relationship of compensatory mutations and strain lineage with RIF resistance.
Resistance mutations detected in rpoB mutations were diverse, located both within and outside the RRDR, and had various impacts on the level of RIF resistance. Among mutations not associated with RIF resistance, four were notable for being located within the RRDR (Thr427Ala [1], Thr427Thr [1], and Arg447Arg [2]). These SNPs, which may result in false-positive predictions of resistance when tested by probe-based methods, such as Hain LPA or GeneXpert MTB/RIF, were detected at an exceedingly low rate (4/1,779 [0.22%]) in our strain population. There have been reports of silent RRDR mutations in the past, typically identified at rates below 1% in each setting for which data are available (10, 32–35).
Resistance-conferring rpoB mutations were detected most commonly in MDR and XDR strains; however, a significant portion (9/53 [17%]) of RIF-resistant strains were found to be RIF monoresistant. The incidence of RIF monoresistance varies considerably by setting, ranging from 0% in some settings to as high as 21.4% in South Africa (19, 36–39). Reported incidences of RIF monoresistance may be underestimated if low-level resistant strains are undetected or were excluded from analysis as a result of using critical-concentration MGIT DST. It is noteworthy that other DST systems, such as the Löwenstein-Jensen (LJ) proportion method, reportedly have fewer discrepancies when it comes to these mutations (40). In the present study, 70% of low-level RIF-resistant strains were also RIF monoresistant. Detecting and counting these strains may drastically affect reported rates of RIF resistance, particularly RIF monoresistance. A clear link between HIV positivity and RIF monoresistance is not supported by our data, although this may be a result of the relatively high rates of low-level RIF monoresistance in this study. Previous reports linking HIV infection with RIF monoresistance almost exclusively describe high-level RIF resistance (21). It is notable that the single patient with high-level RIF monoresistance for which HIV status was available was HIV positive. The conclusions we can draw from these data are limited by our small sample size. Although rpoB mutation has been used as a presumptive positive identification of MDR-TB in some settings, our findings suggest that this approach overestimates the rates of additional/multidrug resistance. Therefore, it may be prudent, at least in low-incidence settings, to be cautious of using rpoB mutation as a proxy for MDR-TB. Resistance to isoniazid and to other first-line drugs should be confirmed before switching to a regimen with more serious side effects and potential drug toxicity (41, 42).
WGS identified several rpoB mutations outside the RRDR, most of which were determined to be low-level RIF resistant by follow-up DST. Three strains, none of which were found to exhibit resistance to any other drugs, contained this type of mutation (Ile59Thr [1] and Ile491Phe [2]), and in each case WGS was the first and only indication that a standard therapy was not appropriate. Strains harboring such mutations are at particular risk for treatment failure, as they may go undetected by both conventional targeted genotypic and phenotypic DST (7, 9, 10). In the absence of WGS or MIC testing, these may have gone completely undetected, potentially leading to treatment failure and the acquisition of further resistance. The remaining low-level resistance mutations detected were located within the RRDR and were found in strains with no other resistance (n = 4) and INH-resistant strains (n = 3). Resistance in strains with mutations within the RRDR may be detected more readily than in cases with mutations located outside the RRDR, yet RIF resistance still will be detected only if a sequencing method that evaluates a large portion of the rpoB gene or MIC testing is performed.
MIC testing was performed on a subset of the test strains, including rpoB wild-type strains and strains with rpoB resistance mutations (associated with both low- and high-level RIF resistance). MICs for strains with high-level resistance and double mutations were >16 μg/ml for all isolates tested. The mutations found in double rpoB mutated strains often have low RIF MICs when found alone, but studies utilizing the Bactec MGIT 960 culture system and the agar proportion method have independently reported finding higher levels of RIF resistance when more than one of these mutations is detected (42–47). Low-level resistance mutations were found to have a much lower range of MICs, 0.25 to 1.0 μg/ml, while the MIC range of rpoB wild-type strains was ≤0.12 to 0.5 μg/ml. The MIC range for high-level RIF-resistant strains was clearly distinct compared to the ranges of low-level resistant and susceptible strains; whether using MIC or genotyping, RIF resistance would be readily identified in all strains with high-level resistance.
In our data set, a MIC cutoff for RIF resistance of 0.5 μg/ml provides better performance, 96.2% sensitivity and 95% specificity, than rpoB resistance mutation detection. Using this cutoff, three strains would be misclassified: one strain with rpoB His445Gln would be considered susceptible, and two rpoB wild-type strains would be considered resistant. Previous reports have described the His445Gln mutation, although it has always been reported in strains with a secondary rpoB mutation (43, 44, 48–51). Based on the present study, the increase in MIC with this mutation alone appears to be quite small, but acquisition of a secondary rpoB mutation may result in a substantial increase in MIC. The two strains with MICs of 0.5 μg/ml and wild-type rpoB sequence would not be classified as resistant by genotype, but they exhibit a MIC that is higher than those of all other rpoB wild-type strains and overlaps the MICs of some low-level RIF-resistant strains. The absence of mutation in rpoB does not rule out the possibility of RIF resistance by another mechanism, but no other loci involved with RIF resistance have been well characterized thus far. These results may also represent the limitations of MIC testing, as interpreting MICs can be subjective and no cutoff perfectly delineates low-level RIF-resistant strains from RIF-susceptible strains.
Low-level resistance mutations in rpoB underscore the value of genotypic methods for diagnosing RIF resistance, particularly methods that interrogate the full-length rpoB gene. Testing algorithms may benefit from the inclusion of a genotypic method or a phenotypic method that detects resistance below the critical concentration of 1 μg/ml to detect all clinically relevant RIF resistance.
In addition to rpoB, whole-genome sequencing provided the ability to screen any other genes of interest for mutations possibly related to RIF resistance, including compensatory mutations in rpoA and rpoC. Excluding phylogenetic SNPs found in both resistant and susceptible isolates, we identified potential compensatory mutations in rpoA, rpoB, and rpoC in strains with rpoB resistance mutations. None of these putative compensatory mutations were detected in RIF-susceptible strains.
Each rpoA mutation detected in RIF-resistant strains has been previously reported to be associated with RIF resistance (52–56). Compensatory mutations in rpoC were found most frequently in strains with rpoB Ser450Leu. This link between compensatory mutations and strains with rpoB Ser450Leu has been well established (23, 25, 54, 57–60). A majority of rpoC mutations identified in our study have been described in previous reports (54, 57–66). Many of these mutations are located between codons 431 and 527, a particular region identified as harboring mutations that may compensate for the fitness cost of rpoB resistance mutations (53).
Major lineages were considered when analyzing the type and prevalence of resistance and compensatory mutations. While all four major lineages are represented in the strains isolated from New York State and New York City TB patients, the respective rates of these lineages vary. Moreover, the rates of RIF resistance among these lineages vary, significantly in some cases. RIF resistance was most prevalent among lineage 2 strains and especially low in lineage 1 strains. The association between lineage 2 and higher rates of drug resistance compared to other lineages found in this study has been previously documented (66–68). Other studies, however, have challenged this finding, which suggests that factors other than genetic background play a more critical role in the acquisition of resistance (69, 70).
RIF-resistant strains, and MDR strains in particular, are reportedly less transmissible than susceptible strains in studies done in both low- and high-incidence settings (71–74). In our population, RIF-resistant strains were less likely than susceptible strains to be assigned to a genomic cluster, using a five-SNP threshold. While notable, this finding is limited by the relatively small number of RIF-resistant cases in New York State. Three strains with RIF resistance each matched one previous strain in our database, with zero, three, and three SNPs (clusters 1, 2, and 3). All of these strains had compensatory mutations in rpoB or rpoC, but a direct link between compensatory mutation and increased transmissibility has been rejected in previous studies with larger sample sizes (55, 65, 74). Investigations into clusters 1 and 2 revealed epidemiological links between the patients, further supporting recent transmission. No direct links between the patients in cluster 3 were established, but both of these patients were linked to an MDR outbreak in New York City. The strain implicated in this outbreak, referred to as strain W, was documented to exhibit high rates of transmission during the 1990s (75). These patients may have been independently infected with this strain during this time of high transmission and subsequently became sick after a period of latency.
In conclusion, routine whole-genome sequencing in a clinical setting is a powerful tool for understanding the prevalence and types of RIF resistance in a population while offering insights into strain background, compensation, and strain relatedness. Diagnosing all types of RIF resistance can be challenging, particularly when genotypic and phenotypic results are discordant. MIC testing is a useful tool for determining the significance of rare or novel rpoB mutations, supplementing routine phenotypic testing. Low-level RIF resistance, which causes diagnostic and treatment challenges, is significant in our population and may be underreported, particularly in strains with no other drug resistance. The present study begins to fill in the gaps of our knowledge regarding the prevalence and spread of RIF-resistant TB in New York.
ACKNOWLEDGMENTS
This research project was partially supported by Cooperative Agreement number 1U60OE000103 (CFDA NO. 93.322) with the Association of Public Health Laboratories and the U.S. Centers for Disease Control and Prevention (CDC).
We also acknowledge the Wadsworth Center Advanced Genomic Technologies core for their support for this testing.
Footnotes
Supplemental material is available online only.
For a commentary on this article, see https://doi.org/10.1128/JCM.02328-20.
Contributor Information
Kimberlee A. Musser, Email: kimberlee.musser@health.ny.gov.
Daniel J. Diekema, University of Iowa College of Medicine
REFERENCES
- 1.Ovchinnikov YA, Monastyrskaya GS, Gubanov VV, Guryev SO, Chertov OY, Modyanov NN, Grinkevich VA, Makarova IA, Marchenko TV, Polovnikova IN, Lipkin VM, Sverdlov ED. 1981. The primary structure of Escherichia coli RNA polymerase: nucleotide sequence of the rpoB gene and amino-acid sequence of the beta-subunit. Eur J Biochem 116:621–629. 10.1111/j.1432-1033.1981.tb05381.x. [DOI] [PubMed] [Google Scholar]
- 2.Jin DJ, Gross CA. 1988. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J Mol Biol 202:45–58. 10.1016/0022-2836(88)90517-7. [DOI] [PubMed] [Google Scholar]
- 3.Telenti A, Imboden P, Marchesi F, Matter L, Schopfer K, Bodmer T, Lowrie D, Colston M, Cole S. 1993. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet 341:647–651. 10.1016/0140-6736(93)90417-F. [DOI] [PubMed] [Google Scholar]
- 4.Goldstein BP. 2014. Resistance to rifampicin: a review. J Antibiot 67:625–630. 10.1038/ja.2014.107. [DOI] [PubMed] [Google Scholar]
- 5.Ohno H, Koga H, Kohno S, Tashiro T, Hara K. 1996. Relationship between rifampin MICs for and rpoB mutations of Mycobacterium tuberculosis strains isolated in Japan. Antimicrob Agents Chemother 40:1053–1056. 10.1128/AAC.40.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramaswamy S, Musser J. 1998. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber Lung Dis 79:3–29. 10.1054/tuld.1998.0002. [DOI] [PubMed] [Google Scholar]
- 7.Siu GKH, Zhang Y, Lau TCK, Lau RWT, Ho P-L, Yew W-W, Tsui SKW, Cheng VCC, Yuen K-Y, Yam W-C. 2011. Mutations outside the rifampicin resistance-determining region associated with rifampicin resistance in Mycobacterium tuberculosis. J Antimicrob Chemother 66:730–733. 10.1093/jac/dkq519. [DOI] [PubMed] [Google Scholar]
- 8.Palomino J, Martin A. 2014. Drug resistance mechanisms in Mycobacterium tuberculosis. Antibiotics 3:317–340. 10.3390/antibiotics3030317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Deun A, Barrera L, Bastian I, Fattorini L, Hoffmann H, Kam KM, Rigouts L, Rüsch-Gerdes S, Wright A. 2009. Mycobacterium tuberculosis strains with highly discordant rifampin susceptibility test results. J Clin Microbiol 47:3501–3506. 10.1128/JCM.01209-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williamson DA, Roberts SA, Bower JE, Vaughan R, Newton S, Lowe O, Lewis CA, Freeman JT. 2012. Clinical failures associated with rpoB mutations in phenotypically occult multidrug-resistant Mycobacterium tuberculosis. Int J Tuber Lung Dis 16:216–220. 10.5588/ijtld.11.0178. [DOI] [PubMed] [Google Scholar]
- 11.Van Deun A, Aung KJM, Bola V, Lebeke R, Hossain MA, Rijk WBD, Rigouts L, Gumusboga A, Torrea G, Jong BCD. 2013. Rifampin drug resistance tests for tuberculosis: challenging the gold standard. J Clin Microbiol 51:2633–2640. 10.1128/JCM.00553-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Al-Mutairi NM, Ahmad S, Mokaddas E, Eldeen HS, Joseph S. 2019. Occurrence of disputed rpoB mutations among Mycobacterium tuberculosis isolates phenotypically susceptible to rifampicin in a country with a low incidence of multidrug-resistant tuberculosis. BMC Infect Dis 19:3. 10.1186/s12879-018-3638-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pang Y, Ruan Y-Z, Zhao J, Chen C, Xu C-H, Su W, Huan S-T, Li R-Z, Zhao Y-L, Chin DP, Wang L-X. 2014. Diagnostic dilemma: treatment outcomes of tuberculosis patients with inconsistent rifampicin susceptibility. Int J Tuber Lung Dis 18:357–362. 10.5588/ijtld.13.0459. [DOI] [PubMed] [Google Scholar]
- 14.Shah NS, Lin SG, Barry PM, Cheng Y-N, Schecter G, Desmond E. 2016. Clinical impact on tuberculosis treatment outcomes of discordance between molecular and growth-based assays for rifampin resistance, California 2003–2013. Open Forum Infect Dis 3:ofw150. 10.1093/ofid/ofw150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.APHL Infectious Diseases. 2019. Issues in Mycobacterium tuberculosis complex (MTBC) drug susceptibility testing: rifampin (RIF). https://www.aphl.org/aboutAPHL/publications/Documents/ID-2019Apr-MTBC-DST-RIF-White-Paper.pdf.
- 16.Jo KW, Lee S, Kang MR, Sung H, Kim MN, Shim TS. 2017. Frequency and type of disupted rpoB mutations in Mycobacterium tuberculosis isolates from South Korea. Tuberc Respir Dis 80:270–276. 10.4046/trd.2017.80.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mvelase NR, Pillay M, Sibanda W, Ngozo JN, Brust JCM, Mlisana KP. 2019. rpoB mutations causing discordant rifampicin susceptibility in Mycobacterium tuberculosis: retrospective analysis of prevalence, phenotypic, genotypic, and treatment outcomes. Open Forum Infect Dis 6:ofz065. 10.1093/ofid/ofz065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gibson J, Donnan E, Eather G. 2018. Management of rifampicin mono-resistant tuberculosis in Queensland, Australia: a retrospective case series. Respirol Case Rep 6:e00366. 10.1002/rcr2.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coovadia YM, Mahomed S, Pillay M, Werner L, Mlisana K. 2013. Rifampicin mono-resistance in Mycobacterium tuberculosis in KwaZulu-Natal, South Africa: a significant phenomenon in a high prevalence TB-HIV region. PLoS One 8:e77712. 10.1371/journal.pone.0077712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.World Health Organization. 2014. Global tuberculosis report, 2014 (WHO/HTM/TB/2015.22). World Health Organization, Geneva, Switzerland. [Google Scholar]
- 21.Ridzon R, Whitney CG, McKenna MT, Taylor JP, Ashkar SH, Nitta AT, Harvey SM, Valway S, Woodley C, Cooksey R, Onorato IM. 1998. Risk factors for rifampin mono-resistant tuberculosis. Am J Respir Crit Care Med 157:1881–1884. 10.1164/ajrccm.157.6.9712009. [DOI] [PubMed] [Google Scholar]
- 22.Huo F, Luo J, Shi J, Zong Z, Jing W, Dong W, Dong L, Ma Y, Liang Q, Shang Y, Huang H, Pang Y. 2018. A 10-year comparative analysis shows that increasing prevalence of rifampin-resistant Mycobacterium tuberculosis in China is associated with the transmission of strains harboring compensatory mutations. Antimicrob Agents Chemother 62:e02303-17. 10.1128/AAC.02303-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meftahi N, Namouchi A, Mhenni B, Brandis G, Hughes D, Mardassi H. 2016. Evidence for the critical role of a secondary site rpoB mutation in the compensatory evolution and successful transmission of an MDR tuberculosis outbreak strain. J Antimicrob Chemother 71:324–332. 10.1093/jac/dkv345. [DOI] [PubMed] [Google Scholar]
- 24.Andersson DI. 2006. The biological cost of mutational antibiotic resistance: any practical conclusions? Curr Opin Microbiol 9:461–465. 10.1016/j.mib.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 25.Brandis G, Hughes D. 2013. Genetic characterization of compensatory evolution in strains carrying rpoB Ser531Leu, the rifampicin resistance mutation most frequently found in clinical isolates. J Antimicrob Chemother 68:2493–2497. 10.1093/jac/dkt224. [DOI] [PubMed] [Google Scholar]
- 26.Talwar A, Tsang CA, Price SF, Pratt RH, Walker WL, Schmit KM, Langer AJ. 2019. Tuberculosis—United States, 2018. MMWR Morb Mortal Wkly Rep 68:257–262. 10.15585/mmwr.mm6811a2.30897076 [DOI] [Google Scholar]
- 27.CLSI. 2018. Susceptibility testing of Mycobacteria, Nocardia, and other aerobic Actinomycetes, 3rd ed. CLSI standard M24. CLSI, Wayne, PA. [PubMed] [Google Scholar]
- 28.Halse TA, Edwards J, Cunningham PL, Wolfgang WJ, Dumas NB, Escuyer VE, Musser KA. 2010. Combined real-time PCR and rpoB gene pyrosequencing for rapid identification of Mycobacterium tuberculosis and determination of rifampin resistance directly in clinical specimens. J Clin Microbiol 48:1182–1188. 10.1128/JCM.02149-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shea J, Halse TA, Lapierre P, Shudt M, Kohlerschmidt D, Van Roey P, Limberger R, Taylor J, Escuyer V, Musser KA. 2017. Comprehensive whole-genome sequencing and reporting of drug resistance profiles on clinical cases of Mycobacterium tuberculosis in New York State. J Clin Microbiol 55:1871–1882. 10.1128/JCM.00298-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Votintseva AA, Pankhurst LJ, Anson LW, Morgan MR, Gascoyne-Binzi D, Walker TM, Quan TP, Wyllie DH, Elias CDO, Wilcox M, Walker AS, Peto TEA, Crook DW. 2015. Mycobacterial DNA extraction for whole-genome sequencing from early positive liquid (MGIT) cultures. J Clin Microbiol 53:1137–1143. 10.1128/JCM.03073-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gagneux S, Small PM. 2007. Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect Dis 7:328–337. 10.1016/S1473-3099(07)70108-1. [DOI] [PubMed] [Google Scholar]
- 32.Alonso M, Palacios JJ, Herranz M, Penedo A, Menéndez Á, Bouza E, Viedma DGD. 2011. Isolation of Mycobacterium tuberculosis strains with a silent mutation in rpoB leading to potential misassignment of resistance category. J Clin Microbiol 49:2688–2690. 10.1128/JCM.00659-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Köser CU, Feuerriegel S, Summers DK, Archer JAC, Niemann S. 2012. Importance of the genetic diversity within the Mycobacterium tuberculosis complex for the development of novel antibiotics and diagnostic tests of drug resistance. Antimicrob Agents Chemother 56:6080–6087. 10.1128/AAC.01641-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mathys V, Vyvere MVD, Droogh ED, Soetaert K, Groenen G. 2014. False-positive rifampicin resistance on Xpert MTB/RIF caused by a silent mutation in the rpoB gene. Int J Tuber Lung Dis 18:1255–1257. 10.5588/ijtld.14.0297. [DOI] [PubMed] [Google Scholar]
- 35.Yue J, Shi W, Xie J, Li Y, Zeng E, Wang H. 2003. Mutations in the rpoB gene of multidrug-resistant Mycobacterium tuberculosis isolates from China. J Clin Microbiol 41:2209–2212. 10.1128/jcm.41.5.2209-2212.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mulenga C, Chonde A, Bwalya IC, Kapata N, Kakungu-Simpungwe M, Docx S, Fissette K, Shamputa IC, Portaels F, Rigouts L. 2010. Low occurrence of tuberculosis drug resistance among pulmonary tuberculosis patients from an urban setting, with a long-running DOTS program in Zambia. Tuberc Res Treat 2010:938178. 10.1155/2010/938178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mvelase NR, Balakrishna Y, Lutchminarain K, Mlisana K. 2019. Evolving rifampicin and isoniazid mono-resistance in a high multidrug-resistant and extensively drug-resistant tuberculosis region: a retrospective data analysis. BMJ Open 9:e031663. 10.1136/bmjopen-2019-031663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Villegas L, Otero L, Sterling TR, Huaman MA, Stuyft PVD, Gotuzzo E, Seas C. 2016. Prevalence, risk factors, and treatment outcomes of isoniazid- and rifampicin-mono-resistant pulmonary tuberculosis in Lima, Peru. PLoS One 11:e0152933. 10.1371/journal.pone.0152933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Villa-Rosas C, Laniado-Laborín R, Oceguera-Palao L. 2015. Primary drug resistance in a region with high burden of tuberculosis. A critical problem. Salud Publica Mex 57:177–179. 10.21149/spm.v57i2.7414. [DOI] [PubMed] [Google Scholar]
- 40.Rigouts L, Gumusboga M, de Rijk WB, Nduwamahoro E, Uwizeye C, de Jong B, Van Deun A. 2013. Rifampin resistance missed in automated liquid culture system for Mycobacterium tuberculosis isolates with specific rpoB mutations. J Clin Microbiol 51:2641–2645. 10.1128/JCM.02741-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rufai SB, Kumar P, Singh A, Prajapati S, Balooni V, Singh S. 2014. Comparison of Xpert MTB/RIF with line probe assay for detection of rifampin-monoresistant Mycobacterium tuberculosis. J Clin Microbiol 52:1846–1852. 10.1128/JCM.03005-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nasiri MJ, Zamani S, Pormohammad A, Feizabadi MM, Aslani HR, Amin M, Halabian R, Imani Fooladi AA. 2018. The reliability of rifampicin resistance as a proxy for multidrug-resistant tuberculosis: a systematic review of studies from Iran. Eur J Clin Microbiol Infect Dis 37:9–14. 10.1007/s10096-017-3079-4. [DOI] [PubMed] [Google Scholar]
- 43.Bodmer T, Zürcher G, Imboden P, Telenti A. 1995. Mutation position and type of substitution in the β-subunit of the RNA polymerase influence in-vitro activity of rifamycins in rifampicin-resistant Mycobacterium tuberculosis. J Antimicrob Chemother 35:345–348. 10.1093/jac/35.2.345. [DOI] [PubMed] [Google Scholar]
- 44.Kapur V, Li LL, Iordanescu S, Hamrick MR, Wanger A, Kreiswirth BN, Musser JM. 1994. Characterization by automated DNA sequencing of mutations in the gene (rpoB) encoding the RNA polymerase beta subunit in rifampin-resistant Mycobacterium tuberculosis strains from New York City and Texas. J Clin Microbiol 32:1095–1098. 10.1128/JCM.32.4.1095-1098.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McAlister AJ, Driscoll J, Metchock B. 2015. DNA sequencing for confirmation of rifampin resistance detected by Cepheid Xpert MTB/RIF assay. J Clin Microbiol 53:1752–1753. 10.1128/JCM.03433-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miotto P, Cabibbe AM, Borroni E, Degano M, Cirillo DM. 2018. Role of disputed mutations in the rpoB gene in interpretation of automated liquid MGIT culture results for rifampin susceptibility testing of Mycobacterium tuberculosis. J Clin Microbiol 56:e01599-17. 10.1128/JCM.01599-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ocheretina O, Shen L, Escuyer VE, Mabou MM, Royal-Mardi G, Collins SE, Pape JW, Fitzgerald DW. 2015. Whole genome sequencing investigation of a tuberculosis outbreak in Port-au-Prince, Haiti, caused by a strain with a "low-level" rpoB mutation L511P—insights into a mechanism of resistance escalation. PLoS One 10:e0129207. 10.1371/journal.pone.0129207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jnawali HN, Hwang SC, Park YK, Kim H, Lee YS, Chung GT, Choe KH, Ryoo S. 2013. Characterization of mutations in multi- and extensive drug resistance among strains of Mycobacterium tuberculosis clinical isolates in Republic of Korea. Diagn Microbiol Infect Dis 76:187–196. 10.1016/j.diagmicrobio.2013.02.035. [DOI] [PubMed] [Google Scholar]
- 49.Ramirez MV, Cowart KC, Campbell PJ, Morlock GP, Sikes D, Winchell JM, Posey JE. 2010. Rapid detection of multidrug-resistant Mycobacterium tuberculosis by use of real-time PCR and high-resolution melt analysis. J Clin Microbiol 48:4003–4009. 10.1128/JCM.00812-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang S, Zhao B, Song Y, Zhou Y, Pang Y, Ou X, Li Q, Xia H, Zhao Y. 2013. Molecular characterization of the rpoB gene mutations of Mycobacterium tuberculosis isolated from China. J Tuber Res 1:1–8. 10.4236/jtr.2013.11001. [DOI] [Google Scholar]
- 51.Yang B, Koga H, Ohno H, Ogawa K, Fukuda M, Hirakata Y, Maesaki S, Tomono K, Tashiro T, Kohno S. 1999. Relationship between antimycobacterial activities of rifampicin, rifabutin and KRM-1648 and rpoB mutations of Mycobacterium tuberculosis. J Antimicrob Chemother 43:613–613. 10.1093/jac/42.5.621. [DOI] [PubMed] [Google Scholar]
- 52.Bainomugisa A, Lavu E, Hiashiri S, Majumdar S, Honjepari A, Moke R, Dakulala P, Hill-Cawthorne GA, Pandey S, Marais BJ, Coulter C, Coin L. 2018. Multi-clonal evolution of multi-drug-resistant/extensively drug-resistant Mycobacterium tuberculosis in a high-prevalence setting of Papua New Guinea for over three decades. Microb Genom 4:e000147. 10.1099/mgen.0.000147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Comas I, Borrell S, Roetzer A, Rose G, Malla B, Kato-Maeda M, Galagan J, Niemann S, Gagneux S. 2011. Whole-genome sequencing of rifampicin-resistant Mycobacterium tuberculosis strains identifies compensatory mutations in RNA polymerase genes. Nat Genet 44:106–110. 10.1038/ng.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Q-J, Jiao W-W, Yin Q-Q, Xu F, Li J-Q, Sun L, Xiao J, Li Y-J, Mokrousov I, Huang H-R, Shen A-D. 2016. Compensatory mutations of rifampin resistance are associated with transmission of multidrug-resistant Mycobacterium tuberculosis Beijing genotype strains in China. Antimicrob Agents Chemother 60:2807–2812. 10.1128/AAC.02358-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu Q, Zuo T, Xu P, Jiang Q, Wu J, Gan M, Yang C, Prakash R, Zhu G, Takiff HE, Gao Q. 2018. Have compensatory mutations facilitated the current epidemic of multidrug-resistant tuberculosis? Emerg Microbes Infect 7:98. 10.1038/s41426-018-0101-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wollenberg KR, Desjardins CA, Zalutskaya A, Slodovnikova V, Oler AJ, Quiñones M, Abeel T, Chapman SB, Tartakovsky M, Gabrielian A, Hoffner S, Skrahin A, Birren BW, Rosenthal A, Skrahina A, Earl AM. 2017. Whole-genome sequencing of Mycobacterium tuberculosis provides insight into the evolution and genetic composition of drug-resistant tuberculosis in Belarus. J Clin Microbiol 55:457–469. 10.1128/JCM.02116-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.San LL, Aye KS, Oo NAT, Shwe MM, Fukushima Y, Gordon SV, Suzuki Y, Nakajima C. 2018. Insight into multidrug-resistant Beijing genotype Mycobacterium tuberculosis isolates in Myanmar. Int J Infect Dis 76:109–119. 10.1016/j.ijid.2018.06.009. [DOI] [PubMed] [Google Scholar]
- 58.Jamieson FB, Guthrie JL, Neemuchwala A, Lastovetska O, Melano RG, Mehaffy C. 2014. Profiling of rpoB mutations and MICs for rifampin and rifabutin in Mycobacterium tuberculosis. J Clin Microbiol 52:2157–2162. 10.1128/JCM.00691-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brandis G, Hughes D. 2018. Mechanisms of fitness cost reduction for rifampicin-resistant strains with deletion or duplication mutations in rpoB. Sci Rep 8:17488. 10.1038/s41598-018-36005-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de Vos M, Muller B, Borrell S, Black PA, Helden PDV, Warren RM, Gagneux S, Victor TC. 2013. Putative compensatory mutations in the rpoC gene of rifampin-resistant Mycobacterium tuberculosis are associated with ongoing transmission. Antimicrob Agents Chemother 57:827–832. 10.1128/AAC.01541-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alyamani EJ, Marcus SA, Ramirez-Busby SM, Hansen C, Rashid J, El-Kholy A, Spalink D, Valafar F, Almehdar HA, Fatani AJ, Khiyami MA, Talaat AM. 2019. Genomic analysis of the emergence of drug-resistant strains of Mycobacterium tuberculosis in the Middle East. Sci Rep 9:4474. 10.1038/s41598-019-41162-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lanzas F, Karakousis PC, Sacchettini JC, Ioerger TR. 2013. Multidrug-resistant tuberculosis in panama is driven by clonal expansion of a multidrug-resistant Mycobacterium tuberculosis strain related to the KZN extensively drug-resistant M. tuberculosis strain from South Africa. J Clin Microbiol 51:3277–3285. 10.1128/JCM.01122-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yun YJ, Lee JS, Yoo JC, Cho E, Park D, Kook Y-H, Lee KH. 2018. Patterns of rpoC mutations in drug-resistant Mycobacterium tuberculosis isolated from patients in South Korea. Tuberc Respir Dis 81:222–227. 10.4046/trd.2017.0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eldholm V, Monteserin J, Rieux A, Lopez B, Sobkowiak B, Ritacco V, Balloux F. 2015. Four decades of transmission of a multidrug-resistant Mycobacterium tuberculosis outbreak strain. Nat Commun 6:7119. 10.1038/ncomms8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vargas AP, Rios AA, Grandjean L, Kirwan DE, Gilman RH, Sheen P, Zimic MJ. 2020. Determination of potentially novel compensatory mutations in rpoC associated with rifampin resistance and rpoB mutations in Mycobacterium tuberculosis Clinical isolates from Peru. Int J Mycobacteriol 9:121–137. 10.4103/ijmy.ijmy_27_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ribeiro SCM, Gomes LL, Amaral EP, Andrade MRM, Almeida FM, Rezende AL, Lanes VR, Carvalho ECQ, Suffys PN, Mokrousov I, Lasunskaia EB. 2014. Mycobacterium tuberculosis strains of the modern sublineage of the Beijing family are more likely to display increased virulence than strains of the ancient sublineage. J Clin Microbiol 52:2615–2624. 10.1128/JCM.00498-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Langlois-Klassen D, Kunimoto D, Saunders LD, Chui L, Boffa J, Menzies D, Long R. 2012. A population-based cohort study of Mycobacterium tuberculosis Beijing strains: an emerging public health threat in an immigrant-receiving country? PLoS One 7:e38431. 10.1371/journal.pone.0038431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Qian L, Abe C, Lin TP, Yu MC, Cho SN, Wang S, Douglas JT. 2002. rpoB genotypes of Mycobacterium tuberculosis Beijing family isolates from East Asian countries. J Clin Microbiol 40:1091–1094. 10.1128/jcm.40.3.1091-1094.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang C, Luo T, Sun G, Qiao K, Sun G, Deriemer K, Mei J, Gao Q. 2012. Mycobacterium tuberculosis Beijing strains favor transmission but not drug resistance in China. Clin Infect Dis 55:1179–1187. 10.1093/cid/cis670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rutaihwa LK, Menardo F, Stucki D, Gygli SM, Ley SD, Malla B, Feldmann J, Borrell S, Beisel C, Middelkoop K, Carter EJ, Diero L, Ballif M, Jugheli L, Reither K, Fenner L, Brites D, Gagneux S. 16 April 2019. Multiple introductions of Mycobacterium tuberculosis lineage 2–Beijing into Africa over centuries. Front Ecol Evol 10.3389/fevo.2019.00112. [DOI] [Google Scholar]
- 71.Ghebremichael S, Groenheit R, Pennhag A, Koivula T, Andersson E, Bruchfeld J, Hoffner S, Romanus V, Källenius G. 2010. Drug resistant Mycobacterium tuberculosis of the Beijing genotype does not spread in Sweden. PLoS One 5:e10893. 10.1371/journal.pone.0010893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grandjean L, Gilman RH, Martin L, Soto E, Castro B, Lopez S, Coronel J, Castillo E, Alarcon V, Lopez V, Miguel AS, Quispe N, Asencios L, Dye C, Moore DAJ. 2015. Transmission of multidrug-resistant and drug-susceptible tuberculosis within households: a prospective cohort study. PLoS Med 12:e1001843. 10.1371/journal.pmed.1001843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Knight GM, Zimic M, Funk S, Gilman RH, Friedland JS, Grandjean L. 2018. The relative fitness of drug resistant Mycobacterium tuberculosis: a modelling study of household transmission in Peru. J R Soc Interface 15:20180025. 10.1098/rsif.2018.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Feng J-Y, Jarlsberg LG, Salcedo K, Rose J, Janes M, Lin S-YG, Osmond DH, Jost KC, Soehnlen MK, Flood J, Graviss EA, Desmond E, Moonan PK, Nahid P, Hopewell PC, Kato-Maeda M. 2017. Clinical and bacteriological characteristics associated with clustering of multidrug-resistant tuberculosis. Int J Tuber Lung Dis 21:766–773. 10.5588/ijtld.16.0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Moss AR, Alland D, Telzak E, Hewlett D, Jr, Sharp V, Chiliade P, LaBombardi V, Kabus D, Hanna B, Palumbo L, Brudney K, Weltman A, Stoeckle K, Chirgwin K, Simberkoff M, Moghazeh S, Eisner W, Lutfey M, Kreiswirth B. 1997. A city-wide outbreak of a multiple-drug-resistant strain of Mycobacterium tuberculosis in New York. Int J Tuber Lung Dis 1:115–121. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequences analyzed in this article have been provided to the Centers for Disease Control and Prevention on a monthly basis to contribute to the National TB Genotyping program (https://www.cdc.gov/tb/programs/genotyping/default.htm) as well as the Relational Sequencing TB Data Platform (ReSeqTB) (https://platform.reseqtb.org/), which catalogs a vast amount of genotypic, phenotypic, and related metadata from M. tuberculosis strains to enable the development of clinically useful, WHO-endorsed in vitro diagnostic assays for rapid drug susceptibility testing.