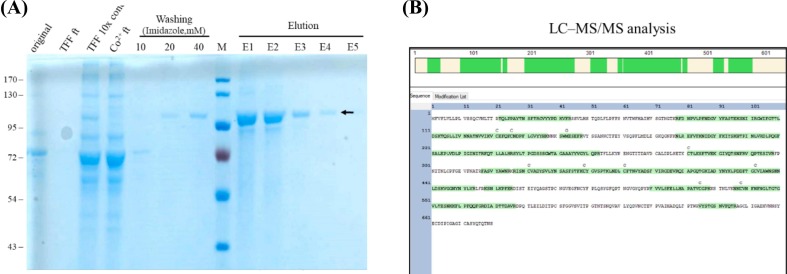
Fig. 1.
Purification and identification of rS1. (A) Western blot analysis to detect rS1 glycoprotein. Protein sizes (kDa) of markers are shown on the left. rS1 is indicated by the arrow. (B) Coomassie gel analysis of a representative purification run of rS1. Proteins were separated on SDS-PAGE and stained with SimplyBlue SafeStain. Original: starting material; TFF flow through (ft): TFF flow-through; TFF 10× con.: 10-fold TFF concentration; Co2+ ft.: flow-through from Co2+ column; Washing: 10, 20, and 40 mM imidazole wash; Elution: elution from HisTrap with 150 and 250 mM imidazole treatments; The respective molecular weights (kDa) of the proteins are indicated on the left. rS1 is indicated by the arrow. (C) Purified rS1 analyzed via mass spectrometry. The amino acid sequences of SARS-CoV-2 S glycoprotein are shown along with the matching peptides obtained from LC–MS/MS analysis (shown in green). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)