CD93 CAR T cells eliminate AML in preclinical models without targeting hematopoietic progenitor cells, and a NOT-gated CAR engineering strategy mitigates on-target, off-tumor toxicity to endothelial cells.
Abstract
Chimeric antigen receptor (CAR) T cells hold promise for the treatment of acute myeloid leukemia (AML), but optimal targets remain to be defined. We demonstrate that CD93 CAR T cells engineered from a novel humanized CD93-specific binder potently kill AML in vitro and in vivo but spare hematopoietic stem and progenitor cells (HSPC). No toxicity is seen in murine models, but CD93 is expressed on human endothelial cells, and CD93 CAR T cells recognize and kill endothelial cell lines. We identify other AML CAR T-cell targets with overlapping expression on endothelial cells, especially in the context of proinflammatory cytokines. To address the challenge of endothelial-specific cross-reactivity, we provide proof of concept for NOT-gated CD93 CAR T cells that circumvent endothelial cell toxicity in a relevant model system. We also identify candidates for combinatorial targeting by profiling the transcriptome of AML and endothelial cells at baseline and after exposure to proinflammatory cytokines.
Significance:
CD93 CAR T cells eliminate AML and spare HSPCs but exert on-target, off-tumor toxicity to endothelial cells. We show coexpression of other AML targets on endothelial cells, introduce a novel NOT-gated strategy to mitigate endothelial toxicity, and demonstrate use of high-dimensional transcriptomic profiling for rational design of combinatorial immunotherapies.
See related commentary by Velasquez and Gottschalk, p. 559.
This article is highlighted in the In This Issue feature, p. 549
Introduction
Acute myeloid leukemia (AML) is the most prevalent acute leukemia in the United States, accounting for more than 11,000 deaths each year, with a 5-year overall survival rate of less than 30% (1). Molecular analysis has accelerated understanding of the genetic diversity of AML and resulted in the approval of numerous molecularly targeted agents, including a Bcl-2 inhibitor (venetoclax), epigenetic modifiers of IDH1/2 (enasidenib, ivosidenib), FLT3 inhibitors (midostaurin, gilteritinib), and a Hedgehog inhibitor (glasdegib; ref. 2). Gemtuzumab ozogamycin, a CD33-directed antibody–drug conjugate, is the only immunotherapy agent currently approved for use in AML (3, 4). Despite these emerging therapies, relapse rates, morbidity, and overall mortality in patients with AML remain high and novel therapies are needed to improve survival and decrease toxicity.
CD19 chimeric antigen receptor (CAR) T-cell therapy has greatly expanded therapeutic options for patients with chemorefractory B-cell acute lymphoblastic leukemia (B-ALL) and diffuse large B-cell lymphoma (DLBCL; refs. 5,6,7,8,9,10,11,12), and these results have prompted intense interest in developing CAR T cells for other hematologic malignancies, including AML. CAR T cells are cytotoxic lymphocytes engineered to express a receptor that combines a tumor antigen recognition domain, normally in the form of a single-chain fragment variable (scFv), with endodomains that impart effector functions derived from the native T-cell receptor and costimulatory proteins. Antitumor efficacy is often accompanied by hematopoietic toxicity due to shared antigen expression within the hematopoietic system (13,14,15,16,17,18), although many have reasoned that hematopoietic toxicity is acceptable if AML CAR T-cell therapy is administered in the context of myeloablation as a bridge to hematopoietic cell transplant (HCT; refs. 19,20,21), or in conjunction with engineering strategies for CAR depletion or regulation (14, 19, 22). Preclinical studies in xenograft models have shown encouraging anti-AML activity of CAR T cells directed against a number of different cell surface molecules (13,14,16,17,18,21, 23,24,25,26,27,28,29,30,31,32,33,34), but only a few have been translated into early-phase clinical trials, most notably CD123 and CD33 (35). The few available published reports of the early clinical experience have shown muted effectiveness in patients compared with the preclinical data (36,37,38,39). Moreover, serious adverse events have been reported. The first two patients on a CD123 CAR trial developed grade 4 capillary leak syndrome, resulting in one patient death and termination of the study (40). Expression of CD123 on endothelial cells may render them susceptible to on-target, off-tumor toxicity (41), a factor that could have contributed to these clinical outcomes. These reports underscore the importance of extending preclinical studies of normal tissue toxicity for new myeloid targets beyond the hematopoietic system.
CD93 is a cell surface lectin that is highly expressed at diagnosis and relapse in a sizable fraction of AML cases (42). In addition, CD93 has been implicated in leukemogenesis and maintenance of a cycling, nonquiescent population of leukemia stem cells in MLL-rearranged (MLLr) AML (43). Here, we engineered a second-generation CAR incorporating a novel CD93-specific scFv, which demonstrated leukemic clearance in preclinical xenograft models and induced limited hematopoietic toxicity. CD93 is largely absent from nonhematopoietic tissues. However, IHC and single-cell RNA sequencing (scRNA-seq) revealed expression in endothelial cells, and we observed targeting of endothelial cell lines with the CD93 CAR. In the same scRNA-seq datasets, CD123 expression mirrored CD93 on endothelial cell subsets, and endothelial cells exposed to inflammatory cytokines expressed both CD123 and CD38, indicating that a risk for endothelial cell toxicity may be significant for AML targets beyond CD93. To address the problem of shared antigen expression between AML and endothelial cells, we employ a NOT-gate strategy and demonstrate mitigation of endothelial cell toxicity by the CD93 CAR. Finally, we implement bulk RNA sequencing (RNA-seq) to more broadly characterize the AML and endothelial surfaceome to provide an inventory of antigens with suitable expression profiles for single or combinatorial targeted AML immunotherapies.
Results
CD93 Is a Novel Cell Surface Target in AML
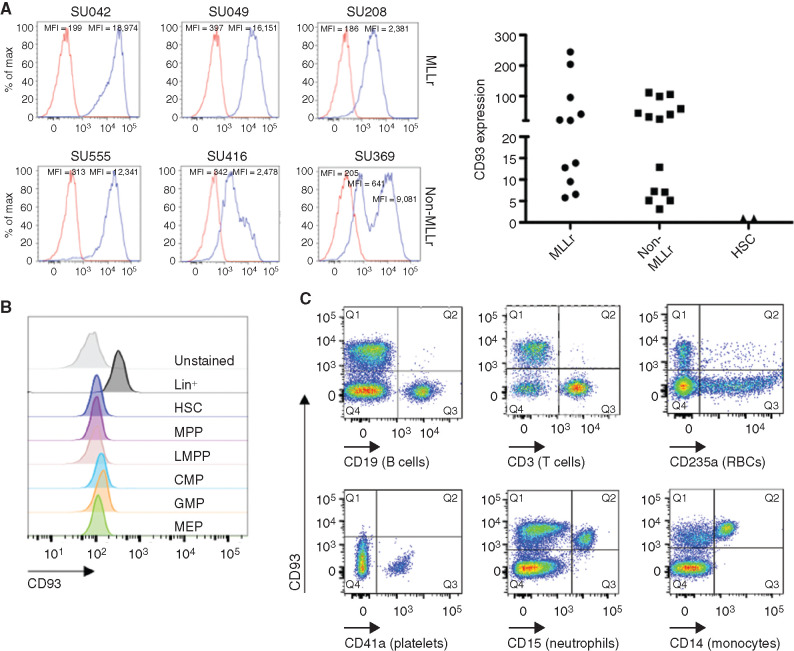
We sought to investigate whether CD93 is a suitable target for CAR-based immunotherapy of human AML. Using flow cytometry, we detected CD93 expression on the majority of primary AML samples, often uniformly and at high levels, including on both MLLr AML and non-MLLr AML (Fig. 1A; Supplementary Fig. S1). CD93 is not expressed on hematopoietic stem cells (HSC) or any myeloid progenitor populations (Fig. 1B) but is expressed on mature myeloid cells, including neutrophils and monocytes. CD93 is absent on lymphocytes, red blood cells, and platelets (Fig. 1C). The absence of expression on hematopoietic progenitors distinguishes CD93 from many previously studied AML targets, including CD33, which is expressed on myeloid progenitors but not on HSCs (44,45,46), and CD123, which is expressed broadly on myeloid and lymphoid progenitors and on HSCs (13, 47, 48).
Figure 1.
CD93 is expressed on AML and mature myeloid cells. A, CD93 expression was evaluated by flow cytometry, in both MLLr or non-MLLr primary AML patient samples, or in HSCs from human healthy bone marrow, identified by Lin−CD34+CD38−CD90+CD45RA− staining. All cells were stained with CD93 (blue) or isotype (red). Left, representative histograms of AML samples. Right, quantification of relative CD93 expression of all AML samples assessed compared with HSCs, represented as fold mean fluorescence intensity (MFI) change compared with isotype control (n = 11 for MLLr, n = 14 for non-MLLr, n = 2 for HSCs). B, Bulk CD34+ selected cells that were either unstained or stained with CD93 and a panel of antibodies to delineate the listed progenitor populations (see Supplementary Fig. S6 for gating strategy). CMP, common myeloid progenitor; GMP, granulocyte–monocyte progenitor; LMPP, lymphoid-primed multipotent progenitor; MEP, megakaryocyte–erythrocyte progenitor; MPP, multipotent progenitor. C, CD93 expression on mature hematopoietic cells was evaluated by staining peripheral blood mononuclear cells (PBMC) with CD93 and lineage markers including CD19 (B cells), CD3 (T cells), CD235a (red blood cells, RBC), CD41a (platelets), CD15 (neutrophils), and CD14 (monocytes). Data are representative of 25 AML samples (A; n = 11 MLLr, n = 14 non-MLLr), 2 healthy donor bone marrow samples (A and B), and 5 healthy donor PBMC samples (C).
CD93 CAR T Cells Mediate Antigen-Specific Effector Function and Cytotoxicity In Vitro against AML Targets
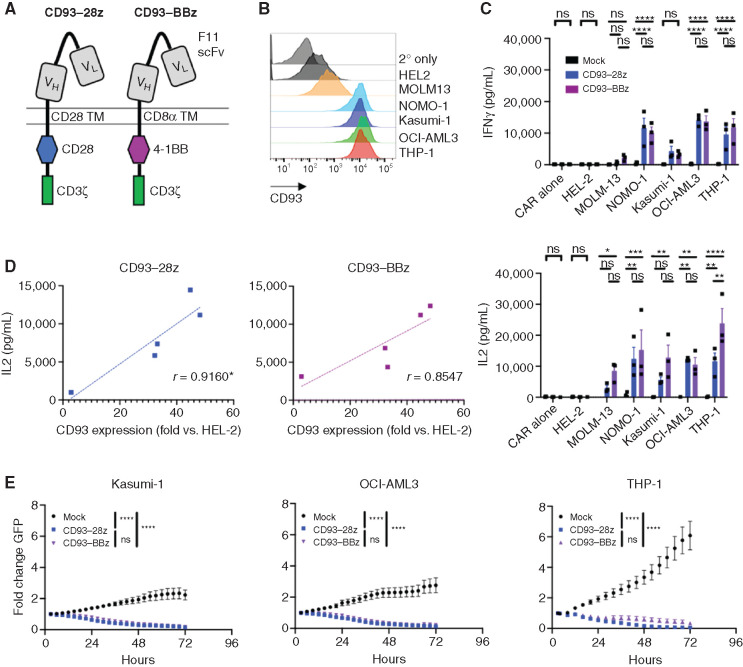
To redirect T-cell specificity against CD93-expressing AML cells, we generated retroviral vectors encoding CD93 CARs that incorporated a CD93-specific scFv derived from a humanized chimeric antibody (F11) developed in our lab (Supplementary Fig. S2). Second-generation CARs were constructed using codon-optimized sequences encoding the F11 scFv at the N-terminus with light and heavy chains connected through a (G3S)4 linker, and fused to either a CD28 hinge-transmembrane, CD28 costimulatory endodomain and CD3ζ (CD93–28z), or to a CD8α hinge-transmembrane, 4-1BB costimulatory endodomain and CD3ζ (CD93–BBz; Fig. 2A). Primary T cells activated and transduced with CD93–28z or CD93–BBz CAR expanded 30- to 50-fold in culture with consistent CAR transduction efficiency of >75% and with comparable mean fluorescence intensity (MFI; Supplementary Fig. S3A–S3C). Similar to previous reports (49), T-cell exhaustion markers PD-1 and TIM-3 were higher in CD93–28z CAR T cells compared with CD93–BBz CAR T cells (Supplementary Fig. S3D).
Figure 2.
CD93 CAR T cells exert antileukemic effects against AML in vitro. A, Structure of CD93–28z and CD93–BBz CARs. From N- to C-terminus, CD93 CARs were designed with VL and VH of the CD93 specific F11 scFv connected via a (G4S)3 linker, CD28 or CD8α TM, CD28 or 4-1BB costimulatory domain, and CD3ζ activation domain. TM, transmembrane. B, CD93 expression on AML cell lines was measured by flow cytometry after staining with biotinylated F11 antibody, followed by streptavidin–APC. C, Mock-transduced, CD93–28z, or CD93–BBz CAR T cells were incubated at a 1:1 effector-to-target (E:T) ratio for 24 hours, and cytokine production was measured in the supernatant by ELISA (unpaired t test, summary data of experiments from three donors). IFNγ secretion: mock versus CD93–28z and CD93–BBz in NOMO-1, OCI-AML3, and THP-1, P < 0.0001. IL2 secretion: mock versus CD93–BBz in MOLM-13, P = 0.0338; mock versus CD93–28z in NOMO-1, P = 0.0027; mock versus CD93–BBz in NOMO-1, P = 0.0002; mock versus CD93–BBz in Kasumi-1, P = 0.0011; mock versus CD93–28z in OCI-AML3, P = 0.0015; mock versus CD93–BBz in OCI-AML3, P = 0.0063; mock versus CD93–28z in THP-1, P = 0.0029; mock versus CD93–BBz in THP-1, P < 0.0001; and CD93–28z versus CD93–BBz in THP-1, P = 0.0017. D, IL2 production of CD93 CAR T cells correlates directly to MFI of CD93 on various AML cell lines, normalized to HEL-2, an AML cell line with low CD93 expression that does not induce cytokine production; *denotes statistical significance: P = 0.0289 for CD93–28z and P = 0.0650 for CD93–BBz (linear regression analysis). E, Mock-transduced, CD93–28z, or CD93–BBz CAR T cells were cocultured with AML cells stably expressing GFP at a 1:1 E:T ratio, and GFP expression was measured in an IncuCyte assay for 72 hours; P < 0.0001 for mock versus CD93 CAR for each cell line (two-way ANOVA, summary data from experiments from n = 2 donors for Kasumi-1 and n = 3 donors for OCI-AML3 and THP-1).
To evaluate CD93 CAR T-cell function in vitro, we analyzed cytokine production and cytotoxicity after coculture with AML cell lines with varying levels of CD93 (Fig. 2B). Of note, the orientation of the light and heavy chain of the scFv did not impact CAR T-cell efficacy in vitro (Supplementary Fig. S4A–S4D). CD93 CAR T cells produced minimal cytokines at baseline but secreted IFNγ and IL2 upon recognition of CD93-expressing AML cells, in contrast to mock-transduced T cells (Fig. 2C). Similar to previous reports emphasizing the importance of target antigen density (50,51,52,53,54,55,56), cytokine production was directly proportional to the intensity of CD93 staining on the surface of AML cells (Fig. 2D; Supplementary Fig. S4E). CD93 CAR T cells also killed AML cells stably expressing GFP in an IncuCyte cytotoxicity assay (Fig. 2E).
CD93 CAR T Cells Exert Potent Antileukemic Effect In Vivo in Cell Lines and Patient-Derived Xenograft Murine Models
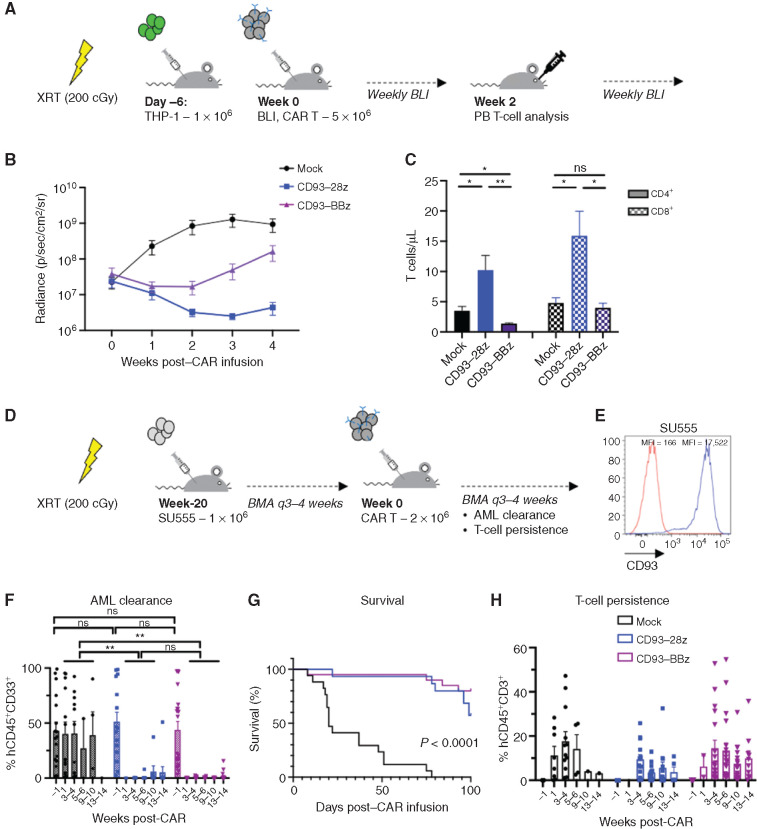
We next evaluated the in vivo efficacy of CD93 CAR T cells in two murine xenograft models of human AML. NOD/SCID/IL2Rγ−/− (NSG) mice were sublethally irradiated and engrafted with luciferase-expressing THP-1 cells. Once engraftment was established by bioluminescent imaging (BLI), the mice were treated with a single dose of mock-transduced, CD93–28z, or CD93–BBz CAR T cells and then monitored by weekly BLI as a surrogate measurement of AML burden (Fig. 3A). Leukemic burden of mice treated with either CD93–28z or CD93–BBz CAR T cells decreased within 1 week of treatment compared with mice treated with mock T cells, a difference that persisted for the duration of the experiment (Fig. 3B; Supplementary Fig. S5A and S5B). We observed increased leukemic clearance in CD93–28z CAR–treated mice, which correlated with greater concentration of CD4+ and CD8+ T cells isolated from peripheral blood at day 14 after CAR T-cell treatment, supporting the importance of in vivo T-cell expansion for leukemic clearance (Fig. 3C).
Figure 3.
CD93 CAR T cells demonstrate antileukemic activity in vivo in murine xenograft models. A, Schematic of the THP-1 xenograft model. NSG mice were sublethally irradiated (200 cGy) on day 7 and then injected via tail vein on day 6 with THP-1 stably expressing GFP/luciferase. BLI was performed on day 0 to quantify engraftment to randomize the treatment groups (n = 5 per group), and then mice were injected via tail vein with mock-transduced, CD93–28z, or CD93–BBz CAR T cells. BLI was measured weekly to quantify AML burden. Peripheral blood was collected at day 14 to measure CAR T-cell expansion. B, Average BLI for THP-1–engrafted mice with leukemic progression in those treated with mock-transduced CAR T cells (black), and leukemic control in those treated with CD93–28z (blue) or CD93–BBz (purple) CAR T cells; P < 0.0001 at weeks 2 and 3 post-CAR (two-way ANOVA, data include experiments with n = 2 donors). C, Measurement of CD4+ and CD8+ T-cell expansion at 2 weeks post–CAR T-cell injection. CD4+ and CD8+ total T cells/μL were measured in the peripheral blood by flow cytometry after red blood cell lysis (unpaired t test). Comparing CD4+ CD93-28z versus CD93-BBz, P = 0.0070; comparing CD8+ CD93-28z versus CD93-BBz, P = 0.0202. D, Schematic of a patient-derived xenograft model. NSG mice were sublethally irradiated (200 cGy) 1 day prior to injection via tail vein of SU555 primary patient-derived AML. Leukemic engraftment was evaluated by BMA every 3 to 4 weeks until 20 weeks after engraftment when AML percentage within the BM was at least 10% on average among the mice in each group randomized to receive either mock-transduced (n = 17), CD93–28z (n = 15), or CD93–BBz (n = 20) CAR T cells. CAR T cells were injected via tail vein, BMA was done 1 week and every 3 to 4 weeks (q3–4 weeks) thereafter, and leukemic clearance and T-cell expansion and persistence were monitored. E, CD93 expression of primary patient AML sample SU555 (blue) compared with isotype (red). F, AML clearance was monitored by flow cytometry of BMA at 1 week and every 3 to 5 weeks after CAR T-cell injection (unpaired t test). G, Survival of mice until 100 days after injection (log-rank Mantel–Cox test). H, T-cell persistence measured from BMA as described in F. Experiments in F–H include summary data from n = 3 donors. PB, peripheral blood; XRT, radiation.
To expand our studies to primary human leukemia samples, we tested CD93 CAR T cells in NSG mice sublethally irradiated and injected with primary AML cells (Fig. 3D), which expressed uniformly high levels of CD93 (Fig. 3E). The mice were monitored for leukemic engraftment by serial bone marrow aspiration (BMA) and treated with mock-transduced or CD93 CAR T cells once human leukemia cells comprised on average at least 10% of the cells isolated from the bone marrow (BM). Of note, engraftment levels at time of treatment were variable and some mice had near complete BM invasion by human AML. Regardless of level of engraftment, all mice treated with CD93 CAR T cells experienced full remission measured by flow cytometry from the BM by 4 weeks and the majority of the mice had no leukemia detectable through 13 weeks after CAR treatment (Fig. 3F). All mice receiving mock-transduced T cells succumbed to disease (Fig. 3G). There was no significant difference among the groups in T-cell persistence at any time point, and all but one CD93 CAR–treated mouse had T-cell persistence through 13 weeks (Fig. 3H). BMA at 1 week after CAR T-cell injection revealed massive cell death in the CD93 CAR–treated groups, precluding analysis of leukemic burden at early time points in these groups (Supplementary Fig. S5C). The mice had minimal systemic toxicity during this time and recovered quickly. A complete blood count 2 weeks after CAR T-cell treatment demonstrated normal hematologic parameters in the CD93 CAR–treated groups compared with leukocytosis, thrombocytopenia, and anemia in the mock-treated group (Supplementary Fig. S5D). One mouse in the CD93–28z- and two in the CD93–BBz-treated groups developed detectable leukemia at later time points, and relapses were driven by CD93-positive cells (Supplementary Fig. S6).
CD93 CAR T Cells Do Not Disrupt Hematopoietic Progenitor Viability or Function
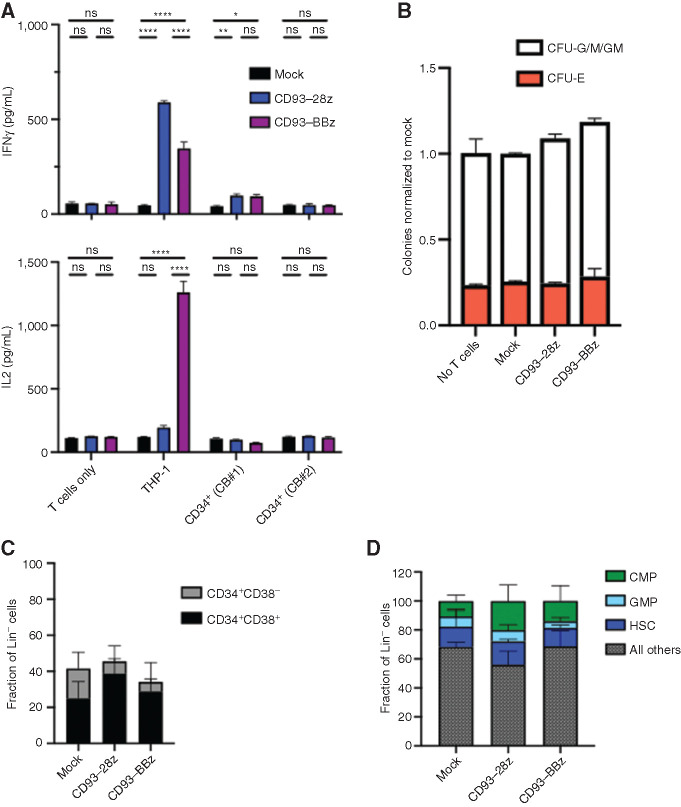
Most AML CAR targets under development are expressed to some extent on normal hematopoietic cells, which is predicted to impact their respective toxicity profiles. Because CD93 expression within the normal hematopoietic compartment is limited to mature myeloid cells (Fig. 1C and D), we hypothesized that CD93 CAR T cells would have minimal impact on hematopoietic stem and progenitor cell (HSPC) viability or function. Indeed, CD93 CAR T cells did not secrete cytokines following exposure to cord blood–derived CD34+ cells, which comprise a variety of HSPC populations (Fig. 4A). To further probe the impact of CD93 CAR T cells on HSPC proliferation and differentiation, cord blood–derived CD34+ cells were untreated or exposed to mock-transduced or CD93 CAR T cells and assayed for colony-forming ability. There was no difference among the erythroid (CFU-E) or granulocyte–macrophage (CFU-G/M/GM) groups (Fig. 4B). Flow cytometric analysis of HSPC subsets after exposure to CD93 CAR T cells confirmed that the proportion of progenitor cells within each subset, whether segregated by CD34+CD38+/CD34+CD38– cells (Fig. 4C) or more specifically by HSCs, common myeloid progenitors, and GM progenitors, was not altered after exposure to CD93 CAR T cells (Fig. 4D; Supplementary Fig. S7). Together these data support that HSPCs are spared from CD93 CAR T-cell toxicity.
Figure 4.
CD93 CAR T cells do not affect hematopoietic progenitor viability or regenerative capacity. A, Mock-transduced, CD93–28z, or CD93–BBz CAR T cells were incubated alone, with THP-1 cells as a positive control, or with two separate cord blood (CB) samples after CD34+ selection. Supernatant was collected at 18 hours, and cytokine production (IFNγ and IL2) was measured by ELISA (unpaired t test, data representative of experiments using n = 2 donors). P < 0.0001 for all differences noted between mock and CD93-28z or CD93-BBz when cocultured with THP-1 cells; P = 0.0073 in mock versus CD93–28z (IFNγ) and P = 0.0133 in mock versus CD93–BBZ (IFNγ) when cocultured with cord blood #1. B, CD34+ cells were sorted after coculture with mock-transduced, CD93–28z, or CD93–BBz CAR T cells, and then suspended in methylcellulose and incubated for 14 days. CFU-G/M/GM and CFU/E colonies from CAR-treated samples were quantified and compared with CD34+ cells not exposed to T cells. Graph represents normalization to mock averaged over two cord blood samples. C and D, After an 18-hour coculture of CAR T cells and cord blood–derived CD34+ cells, flow cytometry was used to quantify the proportion of CD34+ cell subsets, either separated by CD38+ (black)/CD38− (gray; C) or proportions of HSCs (dark blue), common myeloid progenitors (CMP; green), or granulocyte–monocyte progenitors (GMP; light blue; gating strategy detailed in Supplementary Fig. S6; D).
CD93 Is Expressed on Endothelial Cells with a Similar Expression Pattern to CD123
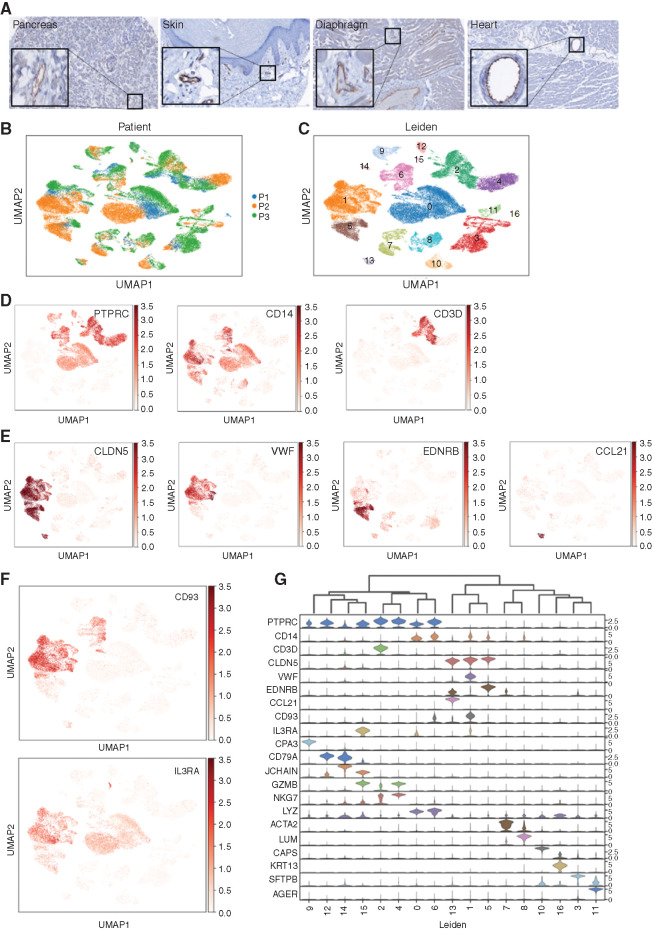
CD93 expression on normal tissue was evaluated by IHC of a tissue microarray. H-scores for all tissues analyzed were <100 (Supplementary Fig. S8A), which is generally accepted as low or no expression (57). Despite low overall expression in normal tissues, strong staining was noted in endothelial cells throughout multiple organ systems (Fig. 5A). Analysis of bulk transcriptional data can mask even substantial expression of a target like CD93 on endothelial cells, which comprise only a fraction of cells within any given organ. Endothelial expression and susceptibility to CAR T-cell targeting have been considered for other AML targets, most notably CD123 (40, 41), so we assessed baseline endothelial expression of CD93 and CD123 by interrogating an scRNA-seq atlas of healthy lung and paired peripheral blood (Fig. 5B; ref. 58). Uniform manifold approximation and projection (UMAP) clustering defined by gene expression profiles resulted in 17 distinct cell populations (Fig. 5C). Hematopoietic cells, defined broadly by PTPRC (CD45), can be found in many clusters, whereas the monocyte marker CD14 and the T-lymphocyte marker CD3D have more restricted expression patterns (Fig. 5D). CLDN5, a pan-endothelial marker (59), is expressed highly in clusters 1, 5, and 13, whereas VWF, EDNRB, and CCL21 likely define stalk-like, tip-like, and high endothelial venule endothelial subsets, respectively (Fig. 5E; refs. 60, 61). CD93 and IL3RA (CD123) are each expressed in a subset of myeloid CD14+ cells as expected but are also clearly expressed in endothelial clusters (Fig. 5F). CD93 and IL3RA expression is compared with the most distinct signature genes for each cell type in a violin plot (Fig. 5G). To ensure the expression patterns are not unique to lung tissue, a similar analysis was performed on scRNA-seq data from nonmalignant pancreatic tissue confirmed clear expression of CD93 and IL3RA within the endothelial compartment despite minimal expression in other clusters (Supplementary Fig. S8B–S8G).
Figure 5.
CD93 and CD123 are expressed on normal myeloid and endothelial cells. A, IHC using CD93 antibody on a tissue microarray with a panel of normal tissues (magnification equivalent to 20×; insets highlight endothelial cell staining). UMAP projection of single cells, colored either by patient sample (B) or Leiden gene expression cluster (C). D, UMAP of single cells, colored by expression of hematopoietic marker genes: PTPRC (CD45) is expressed primarily within clusters 0, 2, 4, 6, 9, 12, and 15; CD14 is expressed primarily within clusters 0, 1, 5, 6, and 8; and CD3D is expressed in cluster 2. E, UMAP projection of single cells, colored by expression of endothelial marker genes marking different endothelial subsets: CLDN5 is expressed in all endothelial clusters (1, 5, and 13); VWF is expressed in stalk-like endothelial cells (cluster 1); EDNRB is expressed in a cluster that contains tip-like endothelial cells (cluster 5); and CCL21 is expressed only in a cluster that represents endothelial venules (cluster 13). F, UMAP of single cells, colored by expression of CD93 and IL3RA (CD123) demonstrated expression in endothelial subclusters (most highly expressed in cluster 1). G, Violin plots displaying the expression level of tissue-specific markers representative of each cluster.
On-Target, Off-Tumor Toxicity of CD93 CAR T Cells against Endothelial Cells Is Mitigated with a NOT-Gated CAR T-cell Strategy In Vitro
Flow cytometry confirmed that CD93 expression on immortalized human umbilical vein endothelial cell (iHUVEC) and telomerase-immortalized human microvascular endothelium cell lines (TIME) was nearly as high as on the AML cell line THP-1 (Fig. 6A). Furthermore, CD93–28z CAR T cells secreted cytokines upon exposure to both iHUVEC and TIME endothelial cell lines (Fig. 6B). Although the scFv contained in the CD93 CAR studied here does not cross-react with murine CD93 and thus endothelial toxicity cannot be assessed in murine models (Supplementary Fig. S9), single-cell transcriptomic analysis of the Tabula Muris database demonstrates a similar expression pattern of CD93 on murine endothelial cells (Supplementary Fig. S10).
Figure 6.
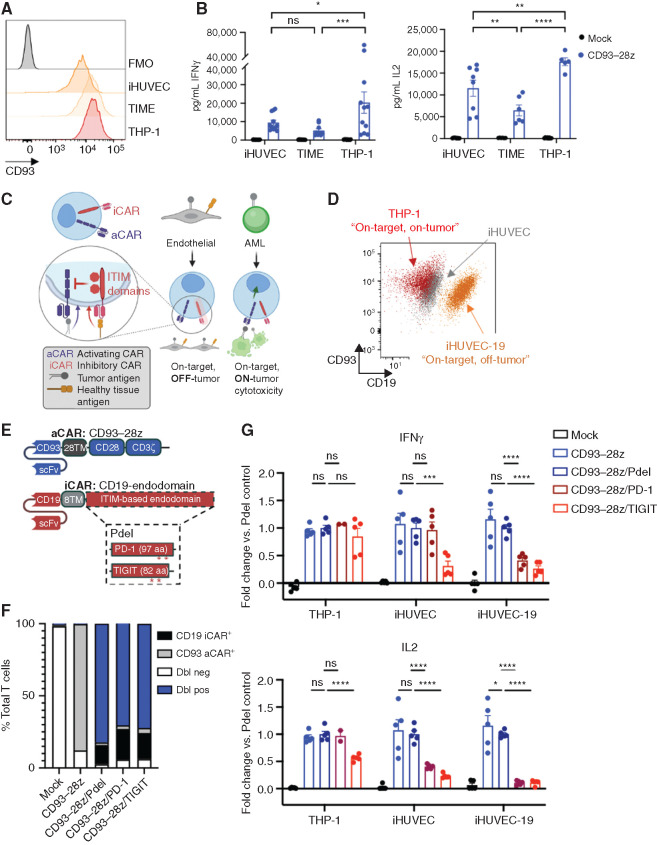
On-target, off-tumor toxicity of CD93 CAR T cells to endothelial cells can be mitigated with an inhibitory CAR (iCAR)–based NOT-gate strategy. A, Flow cytometric analysis of CD93 expression on endothelial cell lines (iHUVEC and TIME) and an AML cell line (THP-1) compared with fluorescence minus one (FMO) negative control. Comparing CD93-28z IFNγ production against cell lines, P = 0.4940 for iHUVEC versus TIME, P = 0.0107 for iHUVEC versus THP-1, and P = 0.0005 for TIME versus THP-1. Comparing CD93-28z IL2 production against cell lines, P = 0.0037 for iHUVEC versus TIME, P = 0.0010 for iHUVEC versus THP-1, and P < 0.0001 for TIME versus THP-1. B, Cytokine secretion by CD93–28z CAR T cells when exposed to endothelial cell lines (iHUVEC and TIME) at a 1:1 effector-to-target ratio. The THP-1 AML cell line is included as a positive control (two-way ANOVA, summary data include experiments from n = 4 donors). C, Schematic of a NOT-gated CAR T-cell system. D, A truncated version of CD19 (extracellular and transmembrane domains) was stably expressed on iHUVECs to generate iHUVEC-19 cells that represent “on-target, off-tumor” cells. Expression of CD93 and CD19 on iHUVEC-19 (orange), iHUVEC (gray), and THP-1 (red) is shown. E, Detailed schematic of a CD93–28z aCAR and an iCAR engineered with a CD19-specific scFv, CD8α transmembrane domain, and an endodomain derived from inhibitory receptors with ITIMs, including PD-1 and TIGIT. A construct with no intracellular signaling domain (Pdel) serves as a control. F, Summary of cotransduction efficiencies of NOT-gated CARs compared with controls. Dbl neg, double negative; Dbl pos, double positive. G, ELISA of supernatant collected after coculture of THP-1, iHUVEC, or iHUVEC-19 with mock-transduced T cells, CD93–28z aCAR T cells only, or NOT-gated CAR T cells; results represented as fold change compared with Pdel control (unpaired t test, summary data include experiments from n = 2 donors). In iHUVEC cell line IFNγ, P = 0.000596 for CD93-28z/Pdel compared to CD93-28z/TIGIT. In iHUVEC-19 for IL2, P = 0.014340 for CD93-28z compared to CD93-28z/Pdel. P < 0.0001 for all other comparisons reaching significance.
Previous groups have demonstrated that on-target, off-tumor toxicity can be overcome with NOT-gated CAR T cells that express a second inhibitory CAR (iCAR) specific for an antigen present on healthy cells but absent on tumor cells. Given the propensity for myeloid and endothelial coexpression of target antigens, we hypothesized that a NOT-gate strategy targeting molecules expressed on endothelium, but not myeloid cells, could overcome this problem. To test this, we created an in vitro model of “on-target, off-tumor” endothelial cells to assess the specificity of iCAR-based NOT-gated CAR T cells (Fig. 6C), wherein the iHUVEC endothelial cell line was engineered to stably express a truncated CD19 (extracellular and transmembrane domain only; Fig. 6D). CD19-specific iCARs were designed to include signaling endodomains from immunoreceptor tyrosine-based inhibitory motif (ITIM)–containing proteins, including PD-1 and TIGIT, or no intracellular signaling domain (Pdel) as a control (Fig. 6E). The NOT-gated CD93 CAR T-cell products exhibited robust expansion, high transduction efficiency, and CD4/8 ratios influenced by donor, not by the presence of the iCAR construct (Fig. 6F; Supplementary Fig. S11A–S11C). Cells that coexpressed iCARs with ITIM-containing endodomains demonstrated decreased expression of exhaustion markers PD-1 and TIM3 at baseline compared with CD93–28z (Supplementary Fig. S11D). Upon coculture with iHUVEC-19 “on-target, off-tumor” cells, NOT-gated CD93 CAR T cells secreted significantly less IFNγ and IL2 compared with CD93–28z/Pdel control. IFNγ production against the “on-target, on-tumor” AML cell line THP-1 was preserved across all constructs. Similar to previous reports (62), the PD-1–based iCAR provided antigen-specific inhibition of cytokine production. The novel TIGIT-based iCAR inhibited cytokine production equally as well in endothelial cells, but this was true upon recognition of both iHUVEC and iHUVEC-19 cells, suggesting more baseline antigen-independent inhibitory signaling compared with the PD-1–based iCAR (Fig. 6G).
Defining Endothelial- and AML-Predominant Expression at Baseline and in an Inflammatory Microenvironment Can Inform Rational Combinatorial CAR T-cell Strategy
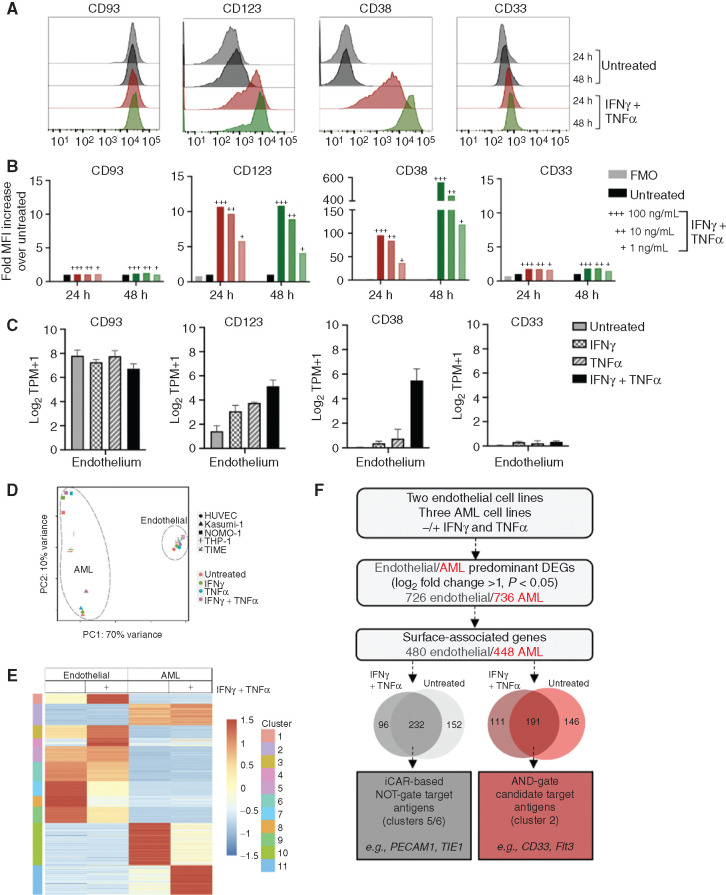
Previous work has demonstrated cytokine inducibility of CD123 expression on endothelial cells that may increase susceptibility to CAR T-cell toxicity (41), but dynamic expression has not been described for other AML targets. Therefore, we performed a targeted expression analysis of CD93 and other select AML targets on endothelial cell lines in the absence and presence of the proinflammatory cytokines IFNγ and TNFα. Cell surface phenotypic analysis revealed that CD123 and CD38 are not expressed highly at baseline on the TIME endothelial cell line, but increase substantially upon exposure to IFNγ and TNFα (Fig. 7A). Cytokine-driven inducible expression is both time and dose dependent (Fig. 7A and B). In contrast, CD33 is not expressed on endothelial cells under any condition. Targeted RNA transcriptional analysis reinforced these expression patterns (Fig. 7C). Pathway analysis suggests that the changes seen in CD123 and CD38 expression are linked by enhancement of the JAK–STAT signaling pathway (Supplementary Fig. S12).
Figure 7.
Endothelial and AML cells exhibit overlap in surface protein expression that is influenced by cytokines. A, TIME cells were either untreated or treated for 24 hours (red) or 48 hours (green) with IFNγ and TNFα (100 ng/mL). CD93 expression patterns on endothelial cells were compared with other common AML targets (CD123, CD38, and CD33) by flow cytometric analysis under both conditions. B, Quantification of endothelial surface protein expression, measured by fold MFI over untreated control. TIME cells were incubated with IFNγ and TNFα at varying doses (+++ = 100 ng/mL, ++ = 10 ng/mL, + = 1 ng/mL) at 24 hours (red) and 48 hours (green), compared with untreated and FMO as negative controls. C, Bulk RNA-seq analysis was performed on two endothelial cell lines (iHUVEC and TIME) that were either untreated or treated with IFNγ, TNFα, or IFNγ and TNFα at 10 ng/mL for 24 hours. Transcriptional levels for selected AML targets on endothelial cells are reported as log2TPM+1. D, RNA was isolated from two endothelial cell lines (iHUVEC and TIME) and three AML cell lines (THP-1, NOMO-1, and Kasumi-1) that were untreated or treated with IFNγ and TNFα (10 ng/mL). E, Heat map of genes that meet criteria for differential expression in AML or endothelial cells reveals clusters of genes with or without exposure to IFNγ and TNFα. Z-scores highlight differences among the cell types and treatment groups. F, Proposed model for gene expression analysis for rational combinatorial targeting (with NOT- or AND-gated CARs) to overcome endothelial toxicity from an AML-directed CAR. DEG, differentially expressed gene.
Figure 6 provides proof of concept that NOT-gated CAR T cells endowed with an iCAR specific for an endothelial cell–specific antigen could overcome on-target, off-tumor toxicity of an AML CAR directed against a target also expressed on endothelial cells. However, the optimal endothelial-specific iCAR target has not been identified. To advance this objective, we implemented an unbiased transcriptional analysis of differential gene expression between endothelial and AML cells, both at baseline and in a proinflammatory microenvironment. To ensure consistency and to avoid a cell line–specific phenomenon, we performed bulk RNA-seq on two endothelial cell lines and three AML cell lines that were either untreated or incubated with cytokines for 24 hours. Principal component analysis (PCA) revealed very little transcriptional variance between the two endothelial cell lines, which formed a grouping distinct from the AML cells. Each AML cell line under all treatment conditions clustered as a group divergent from the others, as would be predicted from their inherent genetic variability (Fig. 7D). When filtering by cell surface designation, genes can be grouped into 11 clusters that emphasize differences between endothelial cells and AML cells, in untreated or in cytokine-treated conditions (Fig. 7E). On the basis of initial filtering parameters, we identified 232 candidate targets for an iCAR-based NOT-gate, which include the well-recognized endothelial cell surface molecules PECAM1 and TIE1 (Fig. 7F). More work will be necessary to validate these as candidate iCAR targets, including verification of preferential endothelial cell surface protein expression compared with AML, confirming lack of expression across a variety of other normal tissues, and identifying available scFv sequences from which to generate iCARs. Ultimately, this strategy could result in an effective and specific NOT-gate platform that could allow for translation of CAR T cells with strong reactivity toward AML-expressed antigens, such as CD93 and CD123, while avoiding endothelial cross-reactivity.
Discussion
Outcomes remain dismal for patients with relapsed or refractory AML despite the emergence of many novel targeted therapies over the last decade. Enthusiasm for adding CAR T cells and other immunotherapies to the AML treatment arsenal is substantial, but progress has been hampered by the challenges of identifying tumor-specific antigens. Here, we identify CD93 as a novel target in AML that may have particular relevance in MLLr AML (43). We report potent antigen-specific, antileukemic activity of CD93 CAR T cells. After demonstrating antileukemic efficacy, we focused our efforts on identifying on-target, off-tumor toxicities. CD93 CAR T cells spare HSPCs, a property distinct from many other AML CARs (13,14,15,16,17,18) but may eliminate mature myeloid cells given CD93 expression on monocytes and neutrophils. The potential for prolonged neutropenia could have serious consequences for patients, and may preclude use of CD93 CAR T cells without a CAR T elimination strategy or as a lead in to HCT. We did not find significant differences in most analyses comparing CD93–28z to CD93–BBz, but if CAR T cells for AML will be used as a bridge to HCT, a CD28 costimulatory domain will likely be desirable because CD28-based CAR T cells generally expand faster and do not persist as long as those with a 4-1BB costimulatory domain (63). This “fast-on, fast-off” strategy could permit stem cell engraftment and hematologic recovery without the need for a CAR ablation strategy. However, further study is needed to understand the balance in patients with AML between immediate CAR T-cell efficacy for leukemic clearance and long-term CAR T persistence for durable leukemic control.
In physiologic and pathologic conditions, blood vessels and blood cells are inextricably linked from early in development. Endothelial and hematopoietic cells share a common precursor, termed a “hemangioblast” (64,65,66). Moreover, HSCs appear to promote migration and maintenance of healthy endothelial cells (67), and endothelial cells secrete signals that increase survival and proliferation of both normal and malignant myeloid cells (68, 69). Emerging data support a developmental trajectory of endothelium to hemogenic endothelium to HSCs and other myeloid progenitors, with significant overlap in essentially a continuum of gene expression (70). Thus, it is not surprising that targets developed for AML will likely often be coexpressed on endothelium, which we have shown here for CD93. Our results also raise concerns about susceptibility of endothelial cells to CAR T cells targeting CD123 and CD38, particularly in the setting of effective CARs that promote a proinflammatory microenvironment that could increase cell surface expression.
With the exception of the serious adverse events described in the UCART123 trial (40), other clinical reports of CD123 CARs have not raised serious safety issues, but this could be falsely reassuring in the absence of reliable potency (38, 39). A patient with relapsed B-ALL received CD38 CAR T cells that resulted in multisystem organ damage refractory to supportive therapy and was highly concerning for on-target, off-tumor effects (71). As more patients are treated with CD123 and CD38 CARs and antileukemic activity improves, patients will need to be monitored for signs of capillary leak syndrome or other endothelial cell toxicity. These reports encourage caution and mandate detailed characterization of on-target, off-tumor toxicity early in CAR development.
Preclinical model systems including in vivo xenografts are insufficient for analysis of on-target, off-tumor toxicity, particularly when the scFv of the CAR does not cross-react with murine tissue, as is the case with the CD93 CAR. Publicly available databases that rely on bulk RNA-seq can be useful for initial screening but may not identify a minority population of susceptible cells. High-dimensional scRNA-seq was recently used to identify a rare population of CD19-expressing mural cells in the brain, which is hypothesized to contribute to neurotoxicity seen with CD19 CAR (72). Similarly, despite low expression on bulk parenchymal tissue, CD93 and CD123 can be clearly identified on endothelial populations. Development of CAR T cells against any novel tumor target requires thorough in silico screening of normal tissues that may have potential for on-target, off-tumor toxicities. This study demonstrates the power of integrating nuanced information provided by scRNA-seq databases as part of these early analyses. However, even thorough analysis of transcriptional data cannot substitute for empiric confirmation of cell surface expression intensity and stability, and demonstration of CAR T-cell safety in relevant model systems. Finally, as a general principle, murine models have not been highly predictive of immune toxicity observed in human studies, emphasizing the importance of carefully controlled early-phase clinical trials for any novel immunotherapeutic agent.
Combinatorial targeting with the goal of circumventing CAR T-cell on-target, off-tumor toxicity against vital normal tissues is an active area of investigation and has been most highly emphasized for the development of CARs targeting solid tumors. The work presented here identifies the importance of developing logic gates for AML as well (73). Autonomous, Boolean logic gate engineering approaches to date have largely focused on AND-gated CARs, which require two antigens to be present on target cells for effective cytotoxicity. Multiple AND-gated CARs have shown promise in preclinical models of AML (74, 75) and other tumors (76,77,78,79). NOT-gated CARs represent an alternative strategy that may prove effective when a particular tissue is the source of shared antigens, such as endothelial cells and myeloid cells, as discussed here. NOT-gates target antigens selectively expressed on the cross-reactive tissue to propagate an inhibitory signal that interferes with the CAR T-cell activation signal (62). Although several groups have proposed NOT-gates as a solution to on-target, off-tumor toxicity of CAR T cells, thus far studies have been largely limited by lack of testing in tissue-specific model systems and by minimal optimization of these platforms (62,80,81,82,83). We provide proof of principle in a relevant in vitro model system that iCAR-based NOT-gated CAR T cells can circumvent CD93 CAR–mediated endothelial cell toxicity. While further characterization and optimization of this platform is necessary, a successful NOT-gated strategy has the potential to expand the breadth of target antigens available for CAR T-cell development and clinical translation.
In addition to other well-defined parameters including tumor heterogeneity, leukemic stem cell expression, and potential for hematopoietic cell toxicity (84, 85), the RNA-seq dataset we have generated could enable antigen selection for single and combinatorial logic-gated AML CARs with a particular focus on avoiding endothelial cell reactivity. For CAR T-cell therapy to become a more viable and widespread option in AML, development of CARs targeting cell surface proteins that have shared expression with some normal tissue seems unavoidable. Endothelial cell expression of AML targets is a particularly important consideration that requires more study and should be assessed for any new AML target.
Methods
Primary Human Samples
AML samples were obtained according to the Administrative Panel on Human Subjects Research Institutional Review Board (IRB)–approved protocols (Stanford IRB numbers 18329 and 6453) after written informed consent. Cord blood was purchased from the New York Blood Center.
Cell Lines
Packaging cell line 293GP, TIME cells, and human AML cell line HEL-2 were purchased from ATCC. MOLM-13, NOMO-1, and OCI-AML3 were purchased from DSMZ. THP-1 cells stably transduced with GFP-luciferase were kindly provided by Dr. Terry Fry at NIH (Bethesda, MD). iHUVECs were a gift from Dr. David Sullivan at Northwestern University (Chicago, IL). iHUVEC-19 cells were generated by transducing iHUVECs with a lentivirus containing a truncated version of CD19, including the extracellular and transmembrane domain only. Transduced cells underwent flow cytometry–based sorting on a BD FACSAria II to isolate a uniform population of CD19+ iHUVECs. All cell lines were verified by short tandem repeat analysis and confirmed to be Mycoplasma negative by PCR.
Generation of CAR Constructs
Generation of Activated CARs.
F11-derived scFvs were codon optimized and custom synthesized by GeneArt (Invitrogen) in either light-heavy or heavy-light orientation and connected with a (G4S)3 linker sequence. The resulting product was subcloned into an MSGV1-based retroviral backbone plasmid encoding the transmembrane and intracellular domains of CD28–CD28 or CD8α–4-1BB and the intracellular domain of CD3ζ, as previously described (49).
Generation of iCAR.
The Pdel iCAR was created by restriction digest deletion of intracellular 4-1BB and CD3ζ intracellular signaling domains of the CD19-BBz CAR that has been described previously (49). PD-1 and TIGIT iCARs were created by subcloning codon-optimized, custom synthesized intracellular domains of PD-1 and TIGIT into the same vector, thereby replacing the 4-1BB and CD3ζ intracellular signaling domains.
Generation of Retroviral Supernatant
Retroviral particles containing CD93–28z and CD93–BBz activating CARs (aCAR), or Pdel, PD-1, or TIGIT iCARs were produced by transfecting 293GP packaging cells with the corresponding CAR plasmid and an RD114 envelope plasmid DNA using Lipofectamine 2000 and harvesting supernatant at 48 and 72 hours after transfection.
Humanized Anti-CD93 Antibody Generation
Monoclonal mouse anti-human CD93 antibodies were generated by immunization of mice using CD93-Fc fusion protein. Hybridomas were generated using standard protocols. Hybridomas were selected, and supernatants from the resulting clones were screened by ELISA and FACS. A mouse hybridoma clone, F11, was identified to produce a mAb with specificity against human CD93. Heavy and light chain variable regions of F11 were cloned from the hybridoma using universal antibody primers. Multiple clones of each V gene product were sequenced to monitor PCR-induced errors. The nucleotide sequences of VH and VL of F11 were determined. To select human antibody frameworks (FR) to be used as templates for compliment determining region (CDR) grafting, the mouse F11 VL and VH regions were compared with those of human germline sequences. Human IGKV2D-29 and IGHV1-2 subgroups were used as the bases for F11 humanization. Amino acid positions in the FR regions that differ between F11 and IGKV2D-29/IGHV1-2 sequences and that may have influence in antigen binding were identified through molecular modeling.
CAR T-cell Transduction
Buffy coats were purchased from Stanford Blood Center under an IRB-exempt protocol, and processed using Lymphoprep density gradient medium and SepMate-50 tubes following manufacturer's instructions. Primary human T cells were positively selected using the RosetteSep Human T cell Enrichment kit (STEMCELL Technologies), and cryopreserved at 1–2 × 107 cells/vial in CryoStor CS10 cryopreservation medium (STEMCELL Technologies). Cryopreserved cells were thawed and activated the same day with CD3/CD28 Dynabeads (Gibco) at a 3:1 bead:cell ratio in T-cell media (RPMI1640 supplemented with 10% FBS, 10 mmol/L HEPES, 2 mmol/L GlutaMAX, 100 U/mL penicillin, 100 μg/mL streptomycin, and 100 IU/mL IL2). Activated T cells were retrovirally transduced with CD93 CAR or cotransduced with CD93 CAR and CD19 iCAR on days 3 and 4 on Retronectin (Takara)-coated plates, and anti-CD3/CD28 beads were removed on day 5. Media and IL2 were changed every 2 to 3 days until day 10 or 11, when T cells were used for assays.
Cytokine Production
0.5–1 × 105 tumor, CD34+, or endothelial cells and CAR T cells at effector-to-target (E:T) ratios between 1:4 and 2:1 were incubated at 37°C in complete RPMI (RPMI1640 supplemented with 10% FBS, 10 mmol/L HEPES, 2 mmol/L GlutaMAX, 100 U/ml penicillin, and 100 μg/mL streptomycin for 18–24 hours in triplicate for each condition). Culture supernatants were collected and analyzed for IFNγ and IL2 by ELISA (BioLegend) as per the manufacturer's instructions.
IncuCyte Lysis Assay
1 × 105 GFP-positive tumor cells and CAR T cells at E:T ratios of 1:8 to 1:1 were cocultured in 200 μL complete RPMI at 37°C for up to 72 hours in triplicate for each condition. Plates were analyzed every 3 hours using the IncuCyte ZOOM Live-Cell Analysis System (Essen Bioscience). Four images per well at 10× zoom were collected at each time point. Total integrated GFP intensity per well was measured. Values were normalized to the starting measurement and plotted over time.
Mice
Immunocompromised NSG mice were purchased from JAX and bred in-house. All mice were bred, housed, and treated under Stanford University Institutional Animal Care and Use Committee (APLAC)–approved protocols. Six- to 8-week-old mice were injected via tail vein with 1 × 106 THP-1 cells stably expressing luciferase or SU555 patient-derived AML cells. Prior to CAR T-cell injection, mice were assigned to groups to equalize pretreatment leukemic burden, either by luminescence values or percentage engraftment. CAR T cells were injected via tail vein at a time and dose provided in the figure legends. In the THP-1 model, leukemia progression was measured by bioluminescence using the IVIS imaging system and analyzed with Living Image software. In the patient-derived xenograft model, leukemia progression was measured by phenotypic analysis of BM aspirates, performed every 3 to 5 weeks on alternating femurs. At least 5 mice per group were treated in each experiment, and each experiment was repeated two to three times as indicated in the figure legend.
Flow Cytometry
All samples were analyzed with an LSR Fortessa or FACSAria II (BD Biosciences), and data were analyzed with FlowJo software. Fc block was used preceding staining of all myeloid cells (Clone 3070, BD Biosciences). CD93 CARs were detected with CD93-Fc chimera protein (R&D Systems), and CD19 iCARs were detected with anti-CD19 CAR idiotype (86) after conjugation to DyLight 650 or Dylight 488, respectively, with an Antibody Labeling Kit (Thermo Fisher Scientific). T-cell phenotype was evaluated with the following antibodies: CD4-BUV395 (Clone SK3, BD Biosciences), CD8-BUV805 (Clone SK1, BD Biosciences), PD-1 (clone eBioJ105, eBioscience), Tim-3-BV510 (clone F38–2E2, BioLegend), LAG-3-PE (clone 3DS223H, eBioscience), and CD39-FITC (Clone A1, BioLegend). CD93 was detected on tumor or endothelial cells using either the F11 antibody or CD93-APC (clone AA4.1, BioLegend). Other antibodies used to phenotype AML cells and endothelial cells included CD45-PerCp/Cy5.5 (clone HI30, eBioscience), CD33-BV421 (clone WM53, BD Biosciences), CD123-BV711 (clone 7G3, BD Biosciences), and CD38-PE (clone HB7, BD Biosciences). Fixable viability stain 780 (BD Biosciences) was used to exclude dead cells from further phenotypic analyses. Antibodies for phenotypic analysis of hematopoietic cells isolated from CD34+ cells included CD45RA-Qdot605 (BioLegend), CD90-FITC (BD Biosciences), CD123-PE (BD Biosciences), CD38-PE-Cy7 (BD Biosciences), CD34-APC (BD Biosciences), and CD10-APC-Cy7 (BioLegend).
Blood Analysis
Peripheral blood samples from the mice were collected on day 14 after CAR T-cell injection to measure CAR T-cell expansion; 50 μL blood was labeled with CD45, CD3, CD4, and CD8, followed by lysis with BD FACS Lysing Solution. Results were quantified using CountBright Absolute Counting Beads (Thermo Fisher) on a BD Fortessa flow cytometer.
Methylcellulose Colony-Forming Assay
Cord blood was obtained from the New York Blood Center with written informed consent according to protocols approved by the Stem Cell Research Oversight Panel (SCRO-291). CD34+ cells were enriched from FICOLL-separated mononuclear cells with the CD34 MicroBead kit for Humans (Miltenyi Biotec, 130–046–703) according to the manufacturer's instructions. After incubation without CAR or with mock-transduced or CD93 CAR T cells for 24 hours, CD34+ cells were sorted on a FACS Aria II (BD Biosciences) and resuspended in 400 μL Iscove's modified Dulbecco's medium with 4 mL MethoCult H4435 (STEMCELL Technologies) according to the manufacturer's instructions and plated in triplicate in 6-well SmartDish (STEMCELL Technologies) plates at 1 × 103 cells per well. Plates were incubated at 37°C and 5% CO2 for 14 days. After 14 days, colonies were scored on an inverted microscope (brand, magnification) for the quantity of erythroid (BFU-E/CFU-E) or CFU-G/M/GM colonies.
IHC and Scoring
CD93 expression was assessed using the Ventana DISCOVERY Ultra autostainer (Ventana Medical Systems). Baked and deparaffinized formalin-fixed, paraffin-embedded tissue sections were incubated in Tris-based buffer (CC1, Ventana) at 95°C for 64 minutes for antigen retrieval, followed by incubation at room temperature for 1 hour with CD93 primary antibody (Clone AA4.1, 1:50, Thermo Fisher) prepared in DISCOVERY Ab Diluent (Ventana). Bound antibodies were visualized with the UltraMap DAB Detection Kit (Ventana). Stained samples were digitized with Leica SCN400 scanner (Leica Microsystems) at magnification equivalent to 20×. The images were subsequently stored in the SlidePath Digital Imaging Hub (Leica Microsystems) at the Vancouver Prostate Centre. For IHC scoring, staining intensity was assigned via a four-point scale system (descriptively, 0 = no staining, 1 = low but detectable degree of staining, 2 = clearly positive cases, and 3 = strong expression) and percentage of positive cells (0%–100%) was also determined. IHC score was then calculated per sample as staining intensity multiplied by percentage of positive cells. The average H-score was calculated for each tissue type.
Analysis of scRNA-seq Data
Processed scRNA-seq data (cell by gene counts table) for human lung were downloaded from Synapse (accession number syn21041850; ref. 58). These data were analyzed using Scanpy v.1.4.3. First, low-quality cells containing fewer than 250 genes, fewer than 500 counts, or more than 25% mitochondrial reads were removed. Counts were depth-normalized to a sum of 10,000 per cell, and then log-transformed with a pseudocount of 1. The top 2,000 variable genes were identified with sc.pp.highly_variable_genes, and effects due to the number of genes detected per cell were regressed out. Data were scaled with max_value = 10, and PCA was performed with default settings. For sc.pp.neighbors, the parameters n_neighbors = 10, n_pcs = 50, random_state = 1 were provided, and Leiden clustering was performed with resolution = 0.1, and random_state = 1. Gene expression was then visualized on the UMAP as the log depth-normalized counts. The sc.tl.rank_genes_group function was used to identify marker genes for each cluster with method = “logreg,” which allowed classification of the endothelial subtypes present in this dataset. For violin plots, gene expression was visualized as depth-normalized counts (not log-transformed). Processed scRNA data for human pancreas were downloaded from the Genome Sequence Archive (project number PRJCA001063; ref. 87). The data were first subset to include only those samples generated from healthy (nontumor) samples, and each sample was processed using Scrublet to remove any potential doublets, with the settings min_counts = 2, min_cells = 3, min_gene_variability_pct1 = 85, n_prin_comps = 30, threshold = 0.2. Data were then processed in Scanpy using the same workflow as above, with the exception of regressing out batch effects due to both the number of genes detected and the individual sample/patient, and using a resolution of 0.15 for Leiden clustering.
Bulk RNA-seq
Bulk RNA was isolated from three AML cell lines (Kasumi-1, NOMO-1, and THP-1) and two endothelial cell lines (iHUVEC and TIME) after 24 hours without treatment or after incubation with IFNγ and/or TNFα at 10 ng/mL. Total RNA was isolated from 2 × 106 cells using Qiagen RNEasy Isolation Kit. Bulk RNA-seq was performed by Novogene using the Illumina Novaseq6000 platform, 150-bp paired-end reads, at 18 to 34 million reads per sample. Reads were quantified using Salmon (v1.2.0; ref. 88) using the human transcriptome (GRCh38.p13, Ensembl release 99) as the reference. Transcript annotation was performed with tximeta (v1.6.2; ref. 89) and differentially expressed genes were identified using DESeq2 (v1.28.1; ref. 90). The variance stabilizing transformation was applied prior to performing PCA with DESeq2. K-means clustering and heat maps were performed with pheatmap (v1.0.12), and Venn diagrams were created with BioVenn.
Gene Set Enrichment Analysis and PSCAN Analysis
Gene set enrichment analysis (GSEA) was performed using GSEA software version 4.0.3 (Broad Institute). The “gene set” permutation type was used, and 1,000 permutations were performed. Gene collections were imported from the Molecular Signatures Database. PSCAN analysis was performed on gene promoters spanning –450 to +50 base pairs flanking the transcriptional start site (91).
Statistical Analysis
Data were analyzed using GraphPad Prism software. Graphs represent individual values or group mean values ± SEM for in vitro and in vivo experiments. P values were calculated with the statistical test described in the corresponding figure legend. P < 0.05 was considered statistically significant, and P values are denoted with asterisks as follows: P > 0.05, not significant (ns); *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Data Sharing Statement
The sequencing datasets generated in this publication have been deposited in NCBI Gene Expression Omnibus (GEO; refs. 92, 93) and are accessible through GEO with the accession number GSE159991.
Authors' Disclosures
R.M. Richards reports a patent for PCT/US2020/020449 issued to Stanford University. K.A. Freitas reports grants from National Science Foundation during the conduct of the study. K.R. Parker reports personal fees and nonfinancial support from Cartography Biosciences outside the submitted work, and is a cofounder and employee of Cartography Biosciences. E. Sotillo reports personal fees from Lyell Immunopharma outside the submitted work. M. Daugaard reports grants and personal fees from Rakovina Therapeutics, VAR2 Pharmaceuticals, and VarCT Diagnostics outside the submitted work. W.-J. Hong reports personal fees from Genentech, Inc. and Imago BioSciences outside the submitted work, as well as a patent for PCT/US2020/020449 pending. H.Y. Chang reports grants from Parker Institute for Cancer Immunotherapy and NIH during the conduct of the study, as well as personal fees from Accent Therapeutics, Boundless Bio, Cartography Bio, 10x Genomics, Arsenal Biosciences, and Spring Discovery outside the submitted work. A.T. Satpathy reports grants from Cancer Research Institute, Parker Institute for Cancer Immunotherapy, and Burroughs Wellcome Fund during the conduct of the study, as well as personal fees from Immunai and Cartography Biosciences outside the submitted work. R.G. Majzner reports personal fees from Syncopation Life Sciences, Lyell Immunopharma, NKarta, Zai Lab, Gamma Delta Therapeutics, Aptorum Group, Illumina Radiopharmaceuticals, and Suda Pharmaceuticals outside the submitted work, as well as a patent for CD93-targeted CAR T cells pending. R. Majeti reports grants from Leukemia & Lymphoma Society during the conduct of the study; personal fees and other support from BeyondSpring Inc., Coherus Biosciences, and Forty Seven Inc., personal fees from Zenshine Pharmaceuticals and Acuta Capital, grants from CircBio Inc., and other support from Kodikaz Therapeutic Solutions and Pheast Therapeutics outside the submitted work; and a patent for CD93 CAR-T for AML pending. C.L. Mackall reports grants from NIH, St. Baldrick's Foundation, Parker Institute for Cancer Immunotherapy, and Ludwig Cancer Research during the conduct of the study; grants, personal fees, and other support from Lyell Immunopharma, personal fees and other support from Syncopation Life Sciences, Vor, Allogene, and Red Tree Capital, and personal fees from Neoimmune Tech, Immatics, GlaxoSmithKline, Bristol Myers Squibb, Apricity Health, Bryologyx, PACT, and Nektar outside the submitted work; and a patent for CD93-targeting CAR-T to treat AML pending. No disclosures were reported by the other authors.
Authors' Contributions
R.M. Richards: Conceptualization, data curation, formal analysis, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. F. Zhao: Formal analysis, investigation, methodology, writing–review and editing. K.A. Freitas: Data curation, validation, investigation, writing–review and editing. K.R. Parker: Data curation, formal analysis, visualization, writing–review and editing. P. Xu: Investigation, writing–review and editing. A. Fan: Data curation, investigation, visualization, writing–review and editing. E. Sotillo: Conceptualization, writing–review and editing. M. Daugaard: Data curation, visualization, writing–review and editing. H.Z. Oo: Data curation, formal analysis, investigation, visualization, writing–review and editing. J. Liu: Resources, data curation, writing–review and editing. W.-J. Hong: Resources, data curation, writing–review and editing. P.H. Sorensen: Supervision, writing–review and editing. H.Y. Chang: Supervision, writing–review and editing. A.T. Satpathy: Supervision, writing–review and editing. R.G. Majzner: Conceptualization, supervision, funding acquisition, writing–review and editing. R. Majeti: Conceptualization, resources, supervision, funding acquisition, writing–review and editing. C.L. Mackall: Conceptualization, resources, supervision, funding acquisition, writing–review and editing.
Supplementary Material
Acknowledgments
This work was supported by the Stanford Maternal Child Health Research Institute, during which time R.M. Richards was the Tashia and John Morgridge Endowed Postdoctoral Fellow, and a St. Baldrick's Foundation Fellowship #604962 (to R.M. Richards). This work was also supported by NIH U54 CA232568–01 (to C.L. Mackall), the Parker Institute for Cancer Immunotherapy (to C.L. Mackall), a Leukemia & Lymphoma Society Translational Research Program Grant 6569–19 (to R. Majeti and C.L. Mackall), the Parker Bridge Scholar Award from the Parker Institute for Cancer Immunotherapy (to A.T. Satpathy), a Technology Impact Award from the Cancer Research Institute (to A.T. Satpathy), a Career Award for Medical Scientists from the Burroughs Wellcome Fund (to A.T. Satpathy), and a SU2C–St. Baldrick's Foundation Pediatric Cancer Dream Team Translational Research Grant (SU2CAACR-DT1113, to C.L. Mackall). Stand Up To Cancer is a division of the Entertainment Industry Foundation. Research grants are administered by the American Association for Cancer Research, the Scientific Partner of SU2C. R.G. Majzner is the Taube Distinguished Scholar for Pediatric Immunotherapy at Stanford University School of Medicine. The authors thank Dr. Evan Weber, Dr. Michael Cleary, and Dr. Masayuki Iwasaki for their critical reading of the manuscript.
Footnotes
Note: Supplementary data for this article are available at Blood Cancer Discovery Online (https://bloodcancerdiscov.aacrjournals.org/).
#R. Majeti and C.L. Mackall contributed equally to this article.
Blood Cancer Discov 2021;2:648–65
References
- 1.Siegel RL, Miller KD, Jemal A.Cancer statistics, 2020. CA Cancer J Clin 2020;70:7–30. [DOI] [PubMed] [Google Scholar]
- 2.Lai C, Doucette K, Norsworthy K.Recent drug approvals for acute myeloid leukemia. J Hematol Oncol 2019;12:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amadori S, Suciu S, Selleslag D, Aversa F, Gaidano G, Musso M, et al. Gemtuzumab ozogamicin versus best supportive care in older patients with newly diagnosed acute myeloid leukemia unsuitable for intensive chemotherapy: results of the randomized phase III EORTC-GIMEMA AML-19 Trial. J Clin Oncol 2016;34:972–9. [DOI] [PubMed] [Google Scholar]
- 4.Castaigne S, Pautas C, Terre C, Raffoux E, Bordessoule D, Bastie JN, et al. Effect of gemtuzumab ozogamicin on survival of adult patients with de-novo acute myeloid leukaemia (ALFA-0701): a randomised, open-label, phase 3 study. Lancet 2012;379:1508–16. [DOI] [PubMed] [Google Scholar]
- 5.Gardner RA, Finney O, Annesley C, Brakke H, Summers C, Leger K, et al. Intent-to-treat leukemia remission by CD19 CAR T cells of defined formulation and dose in children and young adults. Blood 2017;129:3322–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee DW, Kochenderfer JN, Stetler-Stevenson M, Cui YK, Delbrook C, Feldman SA, et al. T cells expressing CD19 chimeric antigen receptors for acute lymphoblastic leukaemia in children and young adults: a phase 1 dose-escalation trial. Lancet 2015;385:517–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maude SL, Frey N, Shaw PA, Aplenc R, Barrett DM, Bunin NJ, et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N Engl J Med 2014;371:1507–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Locke FL, Ghobadi A, Jacobson CA, Miklos DB, Lekakis LJ, Oluwole OO, et al. Long-term safety and activity of axicabtagene ciloleucel in refractory large B-cell lymphoma (ZUMA-1): a single-arm, multicentre, phase 1–2 trial. Lancet Oncol 2019;20:31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schuster SJ, Bishop MR, Tam CS, Waller EK, Borchmann P, McGuirk JP, et al. Tisagenlecleucel in adult relapsed or refractory diffuse large B-cell lymphoma. N Engl J Med 2019;380:45–56. [DOI] [PubMed] [Google Scholar]
- 10.Porter DL, Levine BL, Kalos M, Bagg A, June CH.Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N Engl J Med 2011;365:725–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brentjens RJ, Davila ML, Riviere I, Park J, Wang X, Cowell LG, et al. CD19-targeted T cells rapidly induce molecular remissions in adults with chemotherapy-refractory acute lymphoblastic leukemia. Sci Transl Med 2013;5:177ra38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turtle CJ, Hanafi LA, Berger C, Gooley TA, Cherian S, Hudecek M, et al. CD19 CAR-T cells of defined CD4+:CD8+ composition in adult B cell ALL patients. J Clin Invest 2016;126:2123–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gill S, Tasian SK, Ruella M, Shestova O, Li Y, Porter DL, et al. Preclinical targeting of human acute myeloid leukemia and myeloablation using chimeric antigen receptor-modified T cells. Blood 2014;123:2343–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kenderian SS, Ruella M, Shestova O, Klichinsky M, Aikawa V, Morrissette JJ, et al. CD33-specific chimeric antigen receptor T cells exhibit potent preclinical activity against human acute myeloid leukemia. Leukemia 2015;29:1637–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drent E, Groen RW, Noort WA, Themeli M, Lammerts van Bueren JJ, Parren PW, et al. Pre-clinical evaluation of CD38 chimeric antigen receptor engineered T cells for the treatment of multiple myeloma. Haematologica 2016;101:616–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laborda E, Mazagova M, Shao S, Wang X, Quirino H, Woods AK, et al. Development of a chimeric antigen receptor targeting C-type lectin-like molecule-1 for human acute myeloid leukemia. Int J Mol Sci 2017;18:2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tashiro H, Sauer T, Shum T, Parikh K, Mamonkin M, Omer B, et al. Treatment of acute myeloid leukemia with T cells expressing chimeric antigen receptors directed to C-type lectin-like molecule 1. Mol Ther 2017;25:2202–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jetani H, Garcia-Cadenas I, Nerreter T, Thomas S, Rydzek J, Meijide JB, et al. CAR T-cells targeting FLT3 have potent activity against FLT3(–)ITD(+) AML and act synergistically with the FLT3-inhibitor crenolanib. Leukemia 2018;32:1168–79. [DOI] [PubMed] [Google Scholar]
- 19.Tasian SK, Kenderian SS, Shen F, Ruella M, Shestova O, Kozlowski M, et al. Optimized depletion of chimeric antigen receptor T cells in murine xenograft models of human acute myeloid leukemia. Blood 2017;129:2395–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim MY, Yu KR, Kenderian SS, Ruella M, Chen S, Shin TH, et al. Genetic inactivation of CD33 in hematopoietic stem cells to enable CAR T cell immunotherapy for acute myeloid leukemia. Cell 2018; 173:1439–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Myburgh R, Kiefer JD, Russkamp NF, Magnani CF, Nunez N, Simonis A, et al. Anti-human CD117 CAR T-cells efficiently eliminate healthy and malignant CD117-expressing hematopoietic cells. Leukemia 2020;34:2688–703. [DOI] [PubMed] [Google Scholar]
- 22.Drent E, Poels R, Mulders MJ, van de Donk N, Themeli M, Lokhorst HM, et al. Feasibility of controlling CD38-CAR T cell activity with a Tet-on inducible CAR design. PLoS One 2018;13:e0197349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Casucci M, Nicolis di Robilant B, Falcone L, Camisa B, Norelli M, Genovese P, et al. CD44v6-targeted T cells mediate potent antitumor effects against acute myeloid leukemia and multiple myeloma. Blood 2013;122:3461–72. [DOI] [PubMed] [Google Scholar]
- 24.Chen L, Mao H, Zhang J, Chu J, Devine S, Caligiuri MA, et al. Targeting FLT3 by chimeric antigen receptor T cells for the treatment of acute myeloid leukemia. Leukemia 2017;31:1830–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gomes-Silva D, Atilla E, Atilla PA, Mo F, Tashiro H, Srinivasan M, et al. CD7 CAR T cells for the therapy of acute myeloid leukemia. Mol Ther 2019;27:272–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.John S, Chen H, Deng M, Gui X, Wu G, Chen W, et al. A novel anti-LILRB4 CAR-T cell for the treatment of monocytic AML. Mol Ther 2018;26:2487–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lynn RC, Poussin M, Kalota A, Feng Y, Low PS, Dimitrov DS, et al. Targeting of folate receptor beta on acute myeloid leukemia blasts with chimeric antigen receptor-expressing T cells. Blood 2015;125:3466–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mardiros A, Dos Santos C, McDonald T, Brown CE, Wang X, Budde LE, et al. T cells expressing CD123-specific chimeric antigen receptors exhibit specific cytolytic effector functions and antitumor effects against human acute myeloid leukemia. Blood 2013;122:3138–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tettamanti S, Marin V, Pizzitola I, Magnani CF, Giordano Attianese GM, Cribioli E, et al. Targeting of acute myeloid leukaemia by cytokine-induced killer cells redirected with a novel CD123-specific chimeric antigen receptor. Br J Haematol 2013;161:389–401. [DOI] [PubMed] [Google Scholar]
- 30.Wang J, Chen S, Xiao W, Li W, Wang L, Yang S, et al. CAR-T cells targeting CLL-1 as an approach to treat acute myeloid leukemia. J Hematol Oncol 2018;11:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, Xu Y, Li S, Liu J, Xing Y, Xing H, et al. Targeting FLT3 in acute myeloid leukemia using ligand-based chimeric antigen receptor-engineered T cells. J Hematol Oncol 2018;11:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Westwood JA, Smyth MJ, Teng MW, Moeller M, Trapani JA, Scott AM, et al. Adoptive transfer of T cells modified with a humanized chimeric receptor gene inhibits growth of Lewis-Y-expressing tumors in mice. Proc Natl Acad Sci U S A 2005;102:19051–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshida T, Mihara K, Takei Y, Yanagihara K, Kubo T, Bhattacharyya J, et al. All-trans retinoic acid enhances cytotoxic effect of T cells with an anti-CD38 chimeric antigen receptor in acute myeloid leukemia. Clin Transl Immunology 2016;5:e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Z, Jiang C, Liu Z, Yang M, Tang X, Wang Y, et al. B7-H3-targeted CAR-T cells exhibit potent antitumor effects on hematologic and solid tumors. Mol Ther Oncolytics 2020;17:180–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Epperly R, Gottschalk S, Velasquez MP.Harnessing T cells to target pediatric acute myeloid leukemia: CARs, BiTEs, and beyond. Children 2020;7:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ritchie DS, Neeson PJ, Khot A, Peinert S, Tai T, Tainton K, et al. Persistence and efficacy of second generation CAR T cell against the LeY antigen in acute myeloid leukemia. Mol Ther 2013;21:2122–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang QS, Wang Y, Lv HY, Han QW, Fan H, Guo B, et al. Treatment of CD33-directed chimeric antigen receptor-modified T cells in one patient with relapsed and refractory acute myeloid leukemia. Mol Ther 2015;23:184–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Budde L, Song JY, Kim Y, Blanchard S, Wagner J, Stein AS, et al. Remissions of acute myeloid leukemia and blastic plasmacytoid dendritic cell neoplasm following treatment with CD123-specific CAR T cells: a first-in-human clinical trial.Blood 2017;130 Suppl 1:811. [Google Scholar]
- 39.Cummins KD, Nelson AM, Schmidt A, Luger S, Isaacs RE, Lacey SF, et al. Treating relapsed / refractory (RR) AML with biodegradable anti-CD123 CAR modified T cells.Blood 2017;130 Suppl1:1359. [Google Scholar]
- 40.Cellectis.Cellectis reports clinical hold of UCART123 studies. Cellectis. 2017 Sep 4. Available from:https://www.cellectis.com/en/press/cellectis-reports-clinical-hold-of-ucart123-studies/.
- 41.Sun Y, Wang S, Zhao L, Zhang B, Chen H.IFN-gamma and TNF-alpha aggravate endothelial damage caused by CD123-targeted CAR T cell. Onco Targets Ther 2019;12:4907–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coustan-Smith E, Song G, Shurtleff S, Yeoh AE, Chng WJ, Chen SP, et al. Universal monitoring of minimal residual disease in acute myeloid leukemia. JCI Insight 2018;3:e98561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iwasaki M, Liedtke M, Gentles AJ, Cleary ML.CD93 marks a non-quiescent human leukemia stem cell population and is required for development of MLL-rearranged acute myeloid leukemia. Cell Stem Cell 2015;17:412–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Griffin JD, Linch D, Sabbath K, Larcom P, Schlossman SF.A monoclonal antibody reactive with normal and leukemic human myeloid progenitor cells. Leuk Res 1984;8:521–34. [DOI] [PubMed] [Google Scholar]
- 45.Andrews RG, Takahashi M, Segal GM, Powell JS, Bernstein ID, Singer JW.The L4F3 antigen is expressed by unipotent and multipotent colony-forming cells but not by their precursors. Blood 1986;68:1030–5. [PubMed] [Google Scholar]
- 46.Robertson MJ, Soiffer RJ, Freedman AS, Rabinowe SL, Anderson KC, Ervin TJ, et al. Human bone marrow depleted of CD33-positive cells mediates delayed but durable reconstitution of hematopoiesis: clinical trial of MY9 monoclonal antibody-purged autografts for the treatment of acute myeloid leukemia. Blood 1992;79:2229–36. [PubMed] [Google Scholar]
- 47.Taussig DC, Pearce DJ, Simpson C, Rohatiner AZ, Lister TA, Kelly G, et al. Hematopoietic stem cells express multiple myeloid markers: implications for the origin and targeted therapy of acute myeloid leukemia. Blood 2005;106:4086–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Testa U, Fossati C, Samoggia P, Masciulli R, Mariani G, Hassan HJ, et al. Expression of growth factor receptors in unilineage differentiation culture of purified hematopoietic progenitors. Blood 1996;88:3391–406. [PubMed] [Google Scholar]
- 49.Long AH, Haso WM, Shern JF, Wanhainen KM, Murgai M, Ingaramo M, et al. 4-1BB costimulation ameliorates T cell exhaustion induced by tonic signaling of chimeric antigen receptors. Nat Med 2015;21:581–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Caruso HG, Hurton LV, Najjar A, Rushworth D, Ang S, Olivares S, et al. Tuning sensitivity of CAR to EGFR density limits recognition of normal tissue while maintaining potent antitumor activity. Cancer Res 2015;75:3505–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fry TJ, Shah NN, Orentas RJ, Stetler-Stevenson M, Yuan CM, Ramakrishna S, et al. CD22-targeted CAR T cells induce remission in B-ALL that is naive or resistant to CD19-targeted CAR immunotherapy. Nat Med 2018;24:20–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu X, Jiang S, Fang C, Yang S, Olalere D, Pequignot EC, et al. Affinity-tuned ErbB2 or EGFR chimeric antigen receptor T cells exhibit an increased therapeutic index against tumors in mice. Cancer Res 2015;75:3596–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramakrishna S, Highfill SL, Walsh Z, Nguyen SM, Lei H, Shern JF, et al. Modulation of target antigen density improves CAR T-cell functionality and persistence. Clin Cancer Res 2019;25:5329–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walker AJ, Majzner RG, Zhang L, Wanhainen K, Long AH, Nguyen SM, et al. Tumor antigen and receptor densities regulate efficacy of a chimeric antigen receptor targeting anaplastic lymphoma kinase. Mol Ther 2017;25:2189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watanabe K, Terakura S, Martens AC, van Meerten T, Uchiyama S, Imai M, et al. Target antigen density governs the efficacy of anti-CD20-CD28-CD3 zeta chimeric antigen receptor-modified effector CD8+ T cells. J Immunol 2015;194:911–20. [DOI] [PubMed] [Google Scholar]
- 56.Majzner RG, Rietberg SP, Sotillo E, Dong R, Vachharajani VT, Labanieh L, et al. Tuning the antigen density requirement for CAR T-cell activity. Cancer Discov 2020;10:702–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hirsch FR, Varella-Garcia M, Bunn PA, Jr, Di Maria MV, Veve R, Bremmes RM, et al. Epidermal growth factor receptor in non-small-cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis. J Clin Oncol 2003;21:3798–807. [DOI] [PubMed] [Google Scholar]
- 58.Travaglini KJ, Nabhan AN, Penland L, Sinha R, Gillich A, Sit RV, et al. A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature 2020;587:619–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morita K, Sasaki H, Furuse M, Tsukita S.Endothelial claudin: claudin-5/TMVCF constitutes tight junction strands in endothelial cells. J Cell Biol 1999;147:185–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ager A, May MJ.Understanding high endothelial venules: lessons for cancer immunology. Oncoimmunology 2015;4:e1008791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen W, Xia P, Wang H, Tu J, Liang X, Zhang X, et al. The endothelial tip-stalk cell selection and shuffling during angiogenesis. J Cell Commun Signal 2019;13:291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fedorov VD, Themeli M, Sadelain M.PD-1- and CTLA-4-based inhibitory chimeric antigen receptors (iCARs) divert off-target immunotherapy responses. Sci Transl Med 2013;5:215ra172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao Z, Condomines M, van der Stegen SJC, Perna F, Kloss CC, Gunset G, et al. Structural design of engineered costimulation determines tumor rejection kinetics and persistence of CAR T cells. Cancer Cell 2015;28:415–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tavian M, Coulombel L, Luton D, Clemente HS, Dieterlen-Lievre F, Peault B.Aorta-associated CD34+ hematopoietic cells in the early human embryo. Blood 1996;87:67–72. [PubMed] [Google Scholar]
- 65.Bailey AS, Jiang S, Afentoulis M, Baumann CI, Schroeder DA, Olson SB, et al. Transplanted adult hematopoietic stems cells differentiate into functional endothelial cells. Blood 2004;103:13–9. [DOI] [PubMed] [Google Scholar]
- 66.Choi K, Kennedy M, Kazarov A, Papadimitriou JC, Keller G.A common precursor for hematopoietic and endothelial cells. Development 1998;125:725–32. [DOI] [PubMed] [Google Scholar]
- 67.Takakura N, Watanabe T, Suenobu S, Yamada Y, Noda T, Ito Y, et al. A role for hematopoietic stem cells in promoting angiogenesis. Cell 2000;102:199–209. [DOI] [PubMed] [Google Scholar]
- 68.Fiedler W, Graeven U, Ergun S, Verago S, Kilic N, Stockschlader M, et al. Vascular endothelial growth factor, a possible paracrine growth factor in human acute myeloid leukemia. Blood 1997;89:1870–5. [PubMed] [Google Scholar]
- 69.Ding L, Saunders TL, Enikolopov G, Morrison SJ.Endothelial and perivascular cells maintain haematopoietic stem cells. Nature 2012;481:457–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guibentif C, Ronn RE, Boiers C, Lang S, Saxena S, Soneji S, et al. Single-cell analysis identifies distinct stages of human endothelial-to-hematopoietic transition. Cell Rep 2017;19:10–9. [DOI] [PubMed] [Google Scholar]
- 71.Guo Y, Feng K, Tong C, Jia H, Liu Y, Wang Y, et al. Efficiency and side effects of anti-CD38 CAR T cells in an adult patient with relapsed B-ALL after failure of bi-specific CD19/CD22 CAR T cell treatment. Cell Mol Immunol 2020;17:430–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Parker KR, Migliorini D, Perkey E, Yost KE, Bhaduri A, Bagga P, et al. Single-cell analyses identify brain mural cells expressing CD19 as potential off-tumor targets for CAR-T immunotherapies. Cell 2020;183:126–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Labanieh L, Majzner RG, Mackall CL.Programming CAR-T cells to kill cancer. Nat Biomed Eng 2018;2:377–91. [DOI] [PubMed] [Google Scholar]
- 74.He X, Feng Z, Ma J, Ling S, Cao Y, Gurung B, et al. Bispecific and split CAR T cells targeting CD13 and TIM3 eradicate acute myeloid leukemia. Blood 2020;135:713–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kugler M, Stein C, Kellner C, Mentz K, Saul D, Schwenkert M, et al. A recombinant trispecific single-chain Fv derivative directed against CD123 and CD33 mediates effective elimination of acute myeloid leukaemia cells by dual targeting. Br J Haematol 2010;150:574–86. [DOI] [PubMed] [Google Scholar]
- 76.Kloss CC, Condomines M, Cartellieri M, Bachmann M, Sadelain M.Combinatorial antigen recognition with balanced signaling promotes selective tumor eradication by engineered T cells. Nat Biotechnol 2013;31:71–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lanitis E, Poussin M, Klattenhoff AW, Song D, Sandaltzopoulos R, June CH, et al. Chimeric antigen receptor T Cells with dissociated signaling domains exhibit focused antitumor activity with reduced potential for toxicity in vivo. Cancer Immunol Res 2013;1:43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen C, Li K, Jiang H, Song F, Gao H, Pan X, et al. Development of T cells carrying two complementary chimeric antigen receptors against glypican-3 and asialoglycoprotein receptor 1 for the treatment of hepatocellular carcinoma. Cancer Immunol Immunother 2017;66:475–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Roybal KT, Rupp LJ, Morsut L, Walker WJ, McNally KA, Park JS, et al. Precision tumor recognition by T cells with combinatorial antigen-sensing circuits. Cell 2016;164:770–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dannenfelser R, Allen GM, VanderSluis B, Koegel AK, Levinson S, Stark SR, et al. Discriminatory power of combinatorial antigen recognition in cancer T cell therapies. Cell Syst 2020;11:215–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Williams JZ, Allen GM, Shah D, Sterin IS, Kim KH, Garcia VP, et al. Precise T cell recognition programs designed by transcriptionally linking multiple receptors. Science 2020;370:1099–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hamburger AE, DiAndreth B, Cui J, Daris ME, Munguia ML, Deshmukh K, et al. Engineered T cells directed at tumors with defined allelic loss. Mol Immunol 2020;128:298–310. [DOI] [PubMed] [Google Scholar]
- 83.Chen Z, Kibler RD, Hunt A, Busch F, Pearl J, Jia M, et al. De novo design of protein logic gates. Science 2020;368:78–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Haubner S, Perna F, Kohnke T, Schmidt C, Berman S, Augsberger C, et al. Coexpression profile of leukemic stem cell markers for combinatorial targeted therapy in AML. Leukemia 2019;33:64–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Perna F, Berman SH, Soni RK, Mansilla-Soto J, Eyquem J, Hamieh M, et al. Integrating proteomics and transcriptomics for systematic combinatorial chimeric antigen receptor therapy of AML. Cancer Cell 2017;32:506–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jena B, Maiti S, Huls H, Singh H, Lee DA, Champlin RE, et al. Chimeric antigen receptor (CAR)-specific monoclonal antibody to detect CD19-specific T cells in clinical trials. PLoS One 2013;8:e57838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Peng J, Sun BF, Chen CY, Zhou JY, Chen YS, Chen H, et al. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res 2019;29:725–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C.Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 2017;14:417–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Love MI, Soneson C, Hickey PF, Johnson LK, Pierce NT, Shepherd L, et al. Tximeta: reference sequence checksums for provenance identification in RNA-seq. PLoS Comput Biol 2020;16:e1007664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Love MI, Huber W, Anders S.Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zambelli F, Pesole G, Pavesi G.Pscan: finding over-represented transcription factor binding site motifs in sequences from co-regulated or co-expressed genes. Nucleic Acids Res 2009;37:W247–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res 2013;41:D991–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Edgar R, Barrett T.NCBI GEO standards and services for microarray data. Nat Biotechnol 2006;24:1471–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.