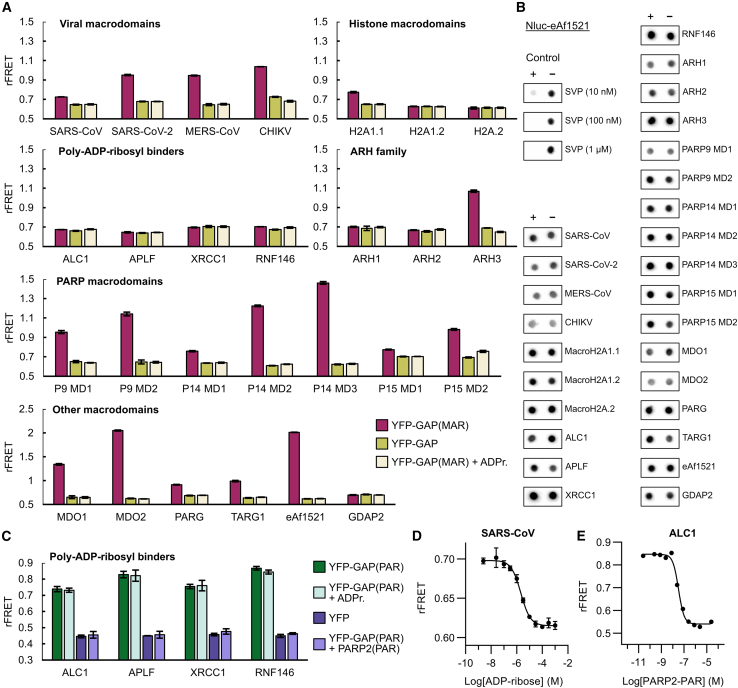
Figure 4.
Testing interactions of reported and potential readers and erasers with YFP-GAP
(A) Interactions of CFP-fused potential and confirmed ADP-ribosyl binders with MARylated YFP-GAP. CFP-fusion proteins (1 μM) were mixed with 5 μM YFP-GAP or with 5 μM YFP-GAP(MAR) in absence or presence of 200 μM ADP-ribose (ADPr.). The ratiometric FRET signals were measured. P9, P14, and P15 denote PARP9, PARP14, and PARP15.
(B) Test of ADP-ribosyl removal from GAP-tag. YFP-GAP(MAR) (10 μM) was prepared in absence (−) or presence (+) of 1 μM CFP-fused proteins or 0.01 μM–1 μM snake venom phosphodiesterase I (SVP). Samples were incubated for 24 h at room temperature and blotted on a nitrocellulose membrane. Detection was done with Nluc-eAf1521.
(C) Interactions of poly-ADP-ribosyl binders with PARylated YFP-GAP. CFP-fusion proteins (250 nM) were mixed with 500 nM YFP or 500 nM YFP-GAP(PAR) in absence or presence of 100 μM ADP-ribose or 2.5 μM automodified PARP2. The ratiometric FRET signals were measured.
(D) Representative dose-response curve of 1 μM CFP-SARS-CoV nsp3 and 5 μM YFP-GAP(MAR) upon competition with ADP-ribose. The control containing no ADP-ribose was set one logarithmic unit below the lowest concentration. Dose-response curves with ADP-ribose for all CFP-fusion proteins are shown in Figure S4.
(E) Representative dose-response curve of 250 nM CFP-ALC1 and 500 nM YFP-GAP(PAR) upon competition with PARylated PARP2. The control containing no PARP2(PAR) was set one logarithmic unit below the lowest concentration, while the control using YFP-GAP(MAR) instead of YFP-GAP(PAR) was set one logarithmic unit above the highest PARP2(PAR) concentration. Data shown for FRET assays are mean ± standard deviation with n = 4 replicates.