Figure 1.
Mouse LOTUS (mLOTUS) expression in neurospheres and differentiated cells derived from 414C2 hiPSC-NS/PCs and gene expression change of LOTUS-expressing NS/PCs in the differentiation marker and neurotrophic factors
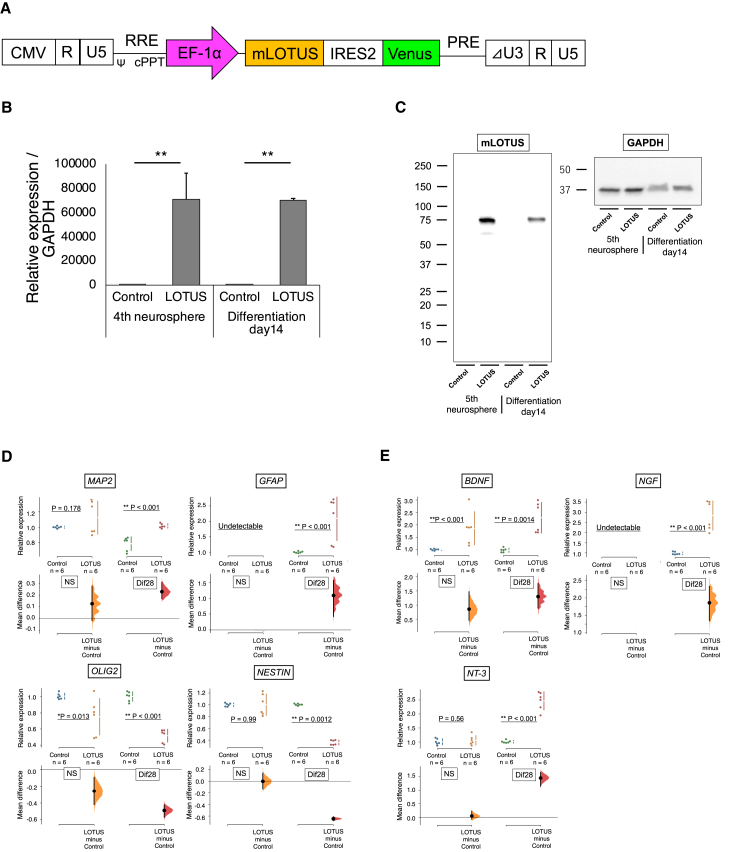
(A) Schematic illustration of the lentiviral vector CSII-EF1α-mLOTUS-IRES-Venus, which expresses mLOTUS cDNA and Venus fluorescent protein gene connected by an internal ribosomal entry site (IRES) under the control of EF1α promoter.
(B) Representative images of the neurosphere derived from control-NS/PCs and LOTUS-NS/PCs visualized by fluorescence microscopy.
(C) Western blot analyses of mLOTUS protein expression in both NS/PCs (LOTUS-NS/PCs; n = 5 independent experiments, control-NS/PCs; n = 5 independent experiments).
(D) Quantitative real-time PCR analyses for the gene expression of MAP2, GFAP, OLIG2, and NESTIN in both NS/PC groups (LOTUS-NS/PCs; n = 6 independent experiments, control-NS/PCs; n = 6 independent experiments).
(E) Quantitative real-time PCR analyses for the gene expression of BDNF, NGF, and NT-3 in both NS/PC groups (LOTUS-NS/PCs; n = 6 independent experiments, control-NS/PCs; n = 6 independent experiments). Values are the mean ± SEM; ∗p < 0.05, ∗∗p < 0.01. Values are the mean ± SEM; ∗p < 0.05, ∗∗p < 0.01. Statistical analysis was performed using the Mann-Whitney U test in each real-time PCR analysis.