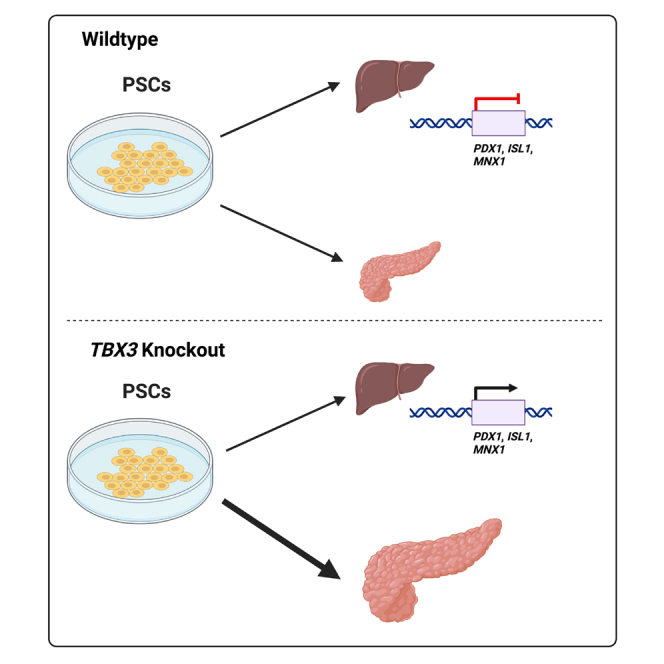
Summary
Tbx3 has been identified as a regulator of liver development in the mouse, but its function in human liver development remains unknown. TBX3 mutant human pluripotent stem cell (PSC) lines were generated using CRISPR/Cas9 genome editing. TBX3 loss led to impaired liver differentiation and an upregulation of pancreatic gene expression, including PDX1, during a hepatocyte differentiation protocol. Other pancreatic genes, including NEUROG3 and NKX2.2, displayed more open chromatin in the TBX3 mutant hepatoblasts. Using a pancreatic differentiation protocol, cells lacking TBX3 generated more pancreatic progenitors and had an enhanced pancreatic gene expression signature at the expense of hepatic gene expression. These data highlight a potential role of TBX3 in regulating hepatic and pancreatic domains during foregut patterning, with implications for enhancing the generation of pancreatic progenitors from PSCs.
Keywords: TBX3, liver, pancreas, development
Graphical abstract
Highlights
-
•
TBX3 null PSCs have impaired hepatocyte differentiation capacity
-
•
TBX3 null hepatocytes have aberrant expression of pancreatic genes, including PDX1
-
•
TBX3 null PSCs have enhanced differentiation capacity into pancreatic progenitors
-
•
Loss of TBX3 leads to increased chromatin accessibility of many pancreatic genes
Mukherjee et al. describe a potential role of TBX3 in patterning hepatic versus pancreatic domains during foregut development. Differentiation of TBX3 null PSCs using a hepatocyte differentiation protocol display impaired hepatocyte gene expression and upregulated expression and chromatin accessibility of pancreatic genes such as PDX1. Using a pancreatic differentiation protocol, TBX3 null PSCs have increased efficiency in pancreatic progenitor generation.
Introduction
The liver and pancreas play vital roles in metabolism and digestion. Both organs arise from the posterior foregut region of the developing gut tube (Deutsch et al., 2001). The gut tube is characterized by an expression pattern of different transcription factors that specify distinct domains for each of the endodermal organs. While the hepatic and pancreatic domains of the gut tube lie in close proximity to each other, they are specified by different signals. Fibroblast growth factor (FGF) from the cardiac mesoderm (Jung et al., 1999) and bone morphogenetic protein 4 from the septum transversum specify the hepatic endoderm (Rossi et al., 2001). Activin and FGF2 signals from the notochord, and retinoic acid from the lateral plate mesoderm specify the dorsal pancreatic endoderm (Hebrok et al., 1998).
T-box transcription factor 3 (TBX3) is involved in development of a number of model systems. Tbx3 drives mesendoderm in Xenopus embryos and mouse pluripotent stem cells (PSCs) (Weidgang et al., 2013), and regulates limb and heart development (Gibson-Brown et al., 1998; Singh et al., 2012). TBX3 is a member of the T-box gene family that acts primarily as a transcriptional repressor (Carlson et al., 2001). In humans, heterozygous mutations in TBX3 result in Ulnar-Mammary Syndrome, a disorder causing defects in limb, mammary, and apocrine gland development (Bamshad et al., 1997). Tbx3 has also been implicated in liver development. Tbx3 is expressed in hepatoblasts, bipotential progenitors that give rise to hepatocytes and cholangiocytes, which comprise the liver bud. Tbx3−/− mice have small liver buds that fail to expand and mature (Lüdtke et al., 2009). Tbx3 has been detected in the mouse pancreas, both during development and in the adult, but its function remains unknown (Begum and Papaioannou, 2011). The role of TBX3 in human liver and pancreas development remains unclear.
PSCs, such as human embryonic stem cells (ESC) and induced PSCs (iPSCs) can give rise to all cell types, and are a model system for studying human development, physiology, and disease. PSCs are easily manipulated using CRISPR/Cas9 genome editing technology, making them valuable for studying the role of specific genes in development and disease (Maguire et al., 2019). Protocols to differentiate PSCs to both hepatocytes (Ogawa et al., 2013; Si-Tayeb et al., 2010) and pancreatic β-cells (D'Amour et al., 2006; Nostro et al., 2011; Rezania et al., 2014) have improved drastically over time. However, it is still difficult to generate functionally mature terminal cell types in vitro.
Here we use PSCs to study the role of TBX3 in human liver and pancreas development. TBX3 mutant PSC lines were generated and differentiated to hepatocytes and pancreatic progenitors. The loss of TBX3 caused a defect in hepatocyte differentiation. Interestingly, the loss of TBX3 resulted in the expression of Pancreatic and Duodenal Homeobox 1 (PDX1), a master regulator for pancreas development (Ahlgren et al., 1996). PSCs lacking TBX3 differentiated more efficiently to pancreatic progenitors than wild-type PSCs. These data suggest that TBX3 may regulate liver development through suppression of pancreatic genes. A better understanding of how TBX3 regulates pancreatic precursor efficiency may provide a potential avenue to improve generation of in vitro–derived pancreatic β-cells for use in therapeutic contexts.
Results and discussion
Loss of TBX3 impairs hepatocyte differentiation in human PSCs
To study the role of TBX3 in human liver development, a TBX3 mutant line was generated in the CHOPi004-A iPSC background (Mukherjee et al., 2020) using CRISPR/Cas9 genome editing technology. Two guide RNAs were designed to generate a 6.9-kb deletion that includes the transcriptional start site and the entire DNA binding domain, generating a nonfunctional protein (Figure 1A). The deletion in both alleles was verified by sequencing and confirmed to have a normal karyotype (Figures S1A and S1B). The genome-edited line is termed iPSC−/−, and the unedited line, designated as iPSC+/+, was used as an isogenic control (Table S1). To confirm the phenotype in a second genetic background, we generated TBX3 mutations in the Mel1 ESC line (Micallef et al., 2012) using the same strategy (Figures S1C and S1D), referred to as ESC−/− (Table S1).
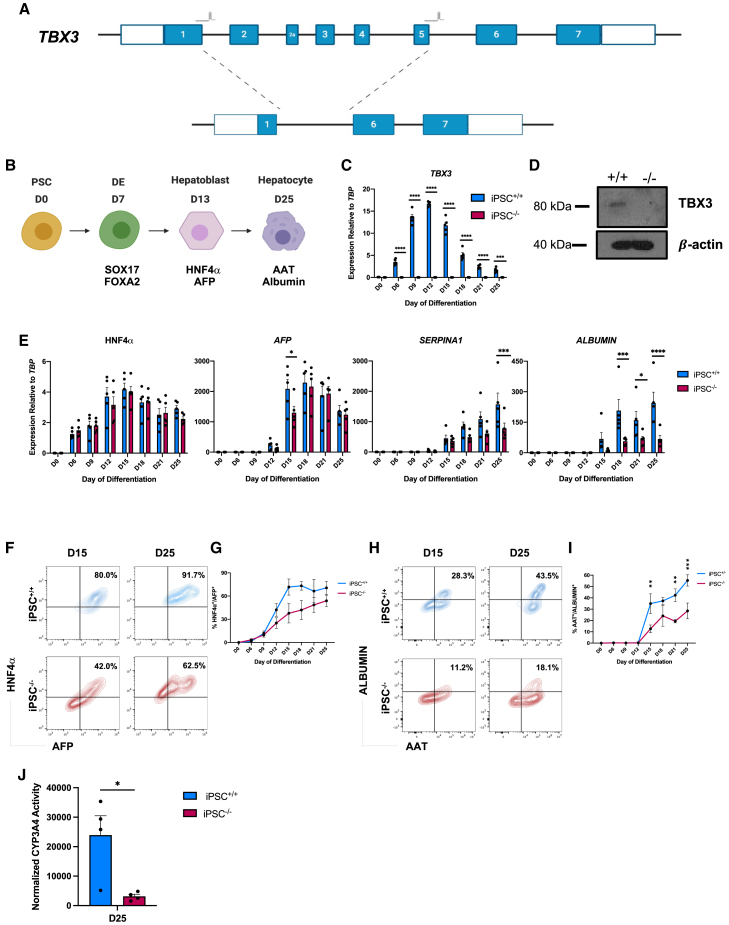
Figure 1.
Loss of TBX3 impairs hepatocyte differentiation
(A) Schematic of endogenous TBX3 locus with sites of gRNAs indicated above exons. Schematic underneath represents TBX3 locus after 6.9-kb deletion.
(B) Schematic of directed differentiation protocol of PSCs to hepatocytes.
(C) Time course of TBX3 expression during hepatocyte differentiation by qRT-PCR (n = 5 separate experiments per time point, per cell line).
(D) Western blot of TBX3 protein in day 12 iPSC+/+ and iPSC−/− hepatoblasts.
(E) Time course of hepatoblast (HNF4α and AFP) and hepatocyte (SERPINA1 and Albumin) markers during hepatocyte differentiation by qRT-PCR (n = 5 per time point, per cell line).
(F) Representative example of HNF4α and AFP expression at day 15 and day 25 by flow cytometry.
(G) Time course of percentage HNF4α+/AFP+ cells by flow cytometry (n = 5 per time point, per cell line).
(H) Representative example of AAT and ALBUMIN expression at day 15 and day 25 by flow cytometry.
(I) Time course of percentage of AAT+/ALBUMIN+ cells in iPSC+/+ and iPSC−/− lines by flow cytometry (n = 5 per time point, per cell line).
(J) Rifampicin-induced CYP3A4 activity in iPSC+/+ and iPSC−/− hepatocytes at day 25 (n = 4 per cell line).
For all statistical analysis, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p ≤ 0.0001.
While TBX3 is expressed in mouse ESCs, it is not expressed in human PSCs (Esmailpour and Huang, 2012), therefore control and mutant lines were differentiated to hepatocytes (Ogawa et al., 2013) to verify loss of TBX3 (Figure 1B). In iPSC+/+ cells, TBX3 expression was induced at the definitive endoderm stage of differentiation and peaked at the hepatoblast stage (day 12). TBX3 was not expressed in the iPSC−/− line during differentiation at the mRNA or protein level (Figures 1C and 1D).
Liver marker expression was measured to determine the impact of TBX3 loss on human liver development. There was no difference in levels of hepatoblast marker HNF4α at the mRNA level (Figure 1E), although expression at the protein level was decreased, along with delayed expression of α-fetoprotein (AFP) in the iPSC−/− line. Tbx3−/− mice develop a normal liver bud but have a small liver due to impaired migration out of the liver bud (Lüdtke et al., 2009), suggesting that Tbx3 is not required for hepatoblast specification, but rather for maturation. This explains the subtle effect observed on hepatoblast marker expression in the differentiation of the iPSC−/− line. A more severe defect was observed later in the differentiation, as expression of hepatocyte markers SERPINA1, which encodes the α1-antitrypsin (AAT) protein, and ALBUMIN were significantly reduced in iPSC−/− cells compared with iPSC+/+ cells (Figure 1E). At the protein level, there were fewer hepatocyte nuclear factor 4 (HNF4)α+/AFP+ cells (Figures 1F and 1G) and AAT+/ALBUMIN+ cells (Figures 1H and 1I) in the iPSC−/− line compared with the iPSC+/+ line. iPSC−/− hepatocytes had significantly reduced CYP3A4 activity compared with iPSC+/+ hepatocytes, indicating that the TBX3 is also needed for hepatocyte functionality (Figure 1J). Similar defects were seen in the ESC−/− line (Figure S2) which were slightly more severe, including a greater downregulation of HNF4α, which may indicate developmental abnormalities are occurring even at the hepatoblast stage. These results indicate that the loss of TBX3 impairs the ability of PSCs to differentiate to hepatocytes, suggesting that TBX3 is important in both mouse and human liver development.
Pancreas-specific genes are upregulated in TBX3 mutant hepatoblasts
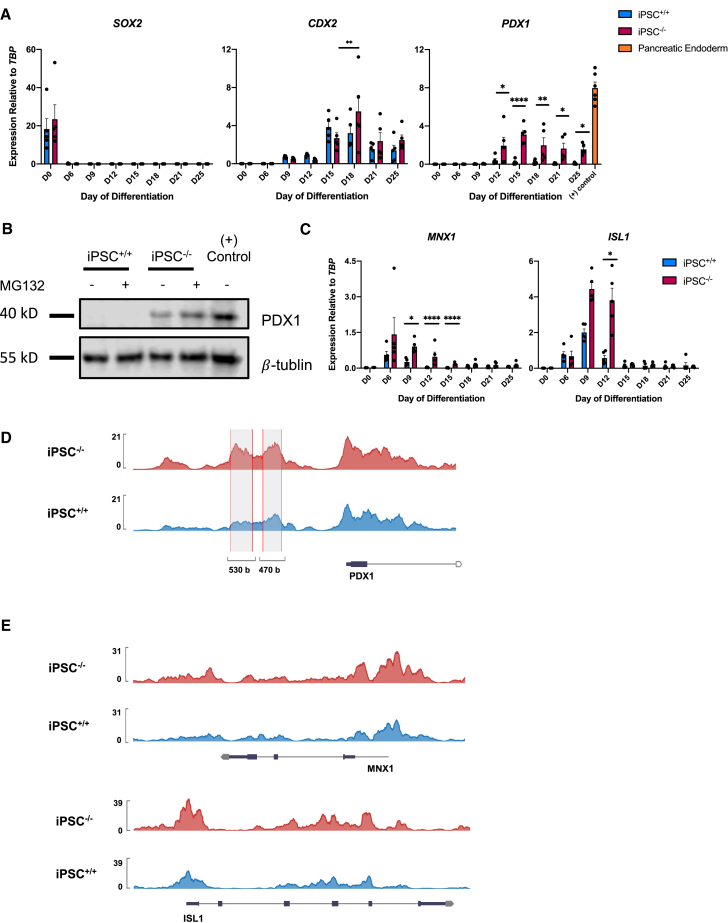
The developing gut tube displays a well-established expression pattern of transcription factors that specify different organ domains (Zorn and Wells, 2009). To determine if the loss of TBX3 impacts gut tube patterning, the expression of the anterior gut tube marker SOX2, posterior gut tube marker CDX2, and pancreatic master regulator PDX1 was examined during hepatocyte differentiation. There was no impact on SOX2 expression, and a minor upregulation of CDX2 in the iPSC−/− line. However, PDX1 was expressed in the iPSC−/− line (Figure 2A), which was surprising, as PDX1 is not expressed during liver differentiation. Because PDX1 protein levels are tightly regulated by ubiquitination and proteasomal degradation (Claiborn et al., 2010), the proteasome inhibitor MG132 (1 μM) was added for 8 h on day 15 when PDX1 expression was highest. In the presence of MG132, iPSC−/− cells had higher PDX1 protein levels than without MG132, while the iPSC+/+ line displayed no PDX1 protein expression with or without MG132 (Figure 2B). This indicates that the loss of TBX3 allows for PDX1 protein expression during the hepatocyte differentiation. In addition, expression of ISL1 (Ahlgren et al., 1997) and Motor Neuron and Pancreas Homeobox 1 (MNX1) (Li et al., 1999), genes involved in early pancreas development, was also increased in the iPSC−/− line (Figure 2C), suggesting a loss of repression of pancreatic genes. Similar results were seen the ESC−/− line (Figures S3A–S3C).
Figure 2.
Pancreatic genes are expressed in the iPSC−/− line during hepatocyte differentiation
(A) Time course of SOX2, PDX1, and CDX2 during hepatocyte differentiation by qRT-PCR (n = 5 per time point, per cell line). PDX1 expression in iPSC+/+ differentiated to pancreatic endoderm for (+) control (n = 6).
(B) Western blot of PDX1 protein in day 15 iPSC+/+ and iPSC−/− immature hepatocytes with or without MG132 and pancreatic endoderm for (+) control.
(C) Time course of early pancreatic markers ISL1 and MNX1 during hepatocyte differentiation by qRT-PCR (n = 5 per time point, per cell line).
(D) Representative example of tracks showing increased accessibility at the PDX1 promoter in iPSC−/− day 12 hepatoblasts.
(E) Representative example of tracks showing differential accessibility at the MNX1 and ISL1 loci in day 12 hepatoblasts. The y axis represents peak height.
For all statistical analysis, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p ≤ 0.0001.
PDX1 drives PSCs to a pancreatic fate during differentiation, in part by repression of hepatic genes, including TBX3 (Teo et al., 2015). The upregulation of PDX1, ISL1, and MNX1 in the iPSC−/− line suggests that TBX3 may be suppressing pancreatic genes to drive hepatocyte differentiation. To understand how the loss of TBX3 impacts chromatin accessibility, we performed assay for transposase-accessible chromatin sequencing (ATAC-seq) in iPSC+/+ and iPSC−/− day 12 hepatoblasts. The PDX1 locus is generally accessible in both control and mutant hepatoblasts, but the promoter region of PDX1 is more accessible in iPSC−/− than in iPSC+/+ hepatoblasts (Figure 2D). This suggests that TBX3 may inhibit PDX1 expression via regulation of the PDX1 promoter, directly or indirectly, to prevent transcription in wild-type hepatoblasts. In addition, the promoter regions of both MNX1 and ISL1 are more accessible in iPSC−/− hepatoblasts than in iPSC+/+ hepatoblasts (Figure 2E). The loss of TBX3 results in inappropriate expression of pancreatic genes during hepatocyte differentiation, suggesting that TBX3 may be important to maintain lineage fidelity of the hepatic domain during foregut patterning.
Loss of TBX3 enhances pancreatic progenitor generation from PSCs
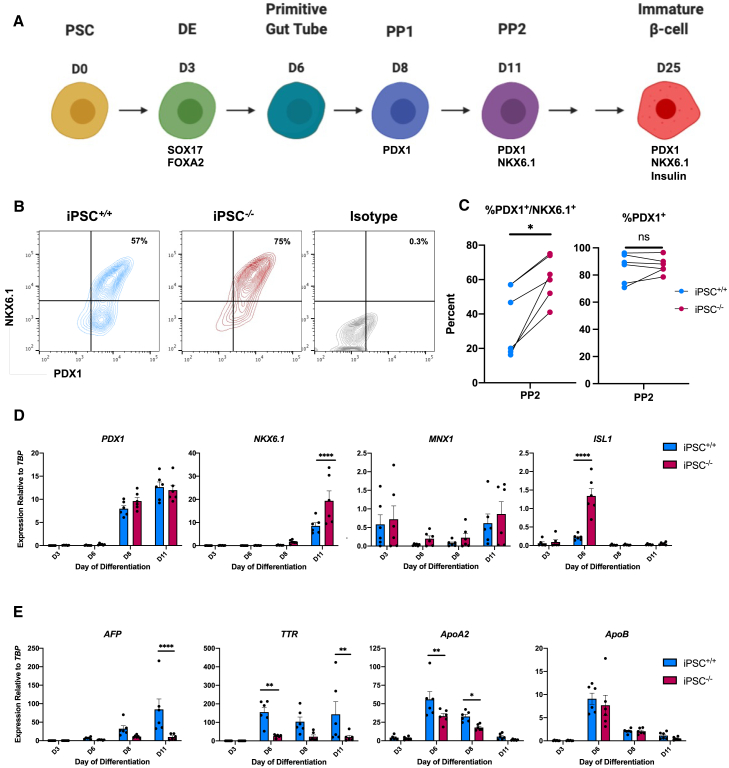
Considering that TBX3 loss led to de-repression of pancreatic genes during liver differentiation, we tested whether loss of TBX3 enhanced pancreatic differentiation. The iPSC+/+ and iPSC−/− lines were differentiated to pancreatic progenitors (Rezania et al., 2014) (Figure 3A). The iPSC−/− line generated a higher percentage of the PDX1+/NKX6.1+ pancreatic progenitor 2 (PP2) population compared with iPSC+/+ cells (Figures 3B and 3C). PDX1 mRNA levels trend higher in the iPSC−/− line at the pancreatic progenitor 1 stage (PP1, day 8) of the pancreas differentiation but do not reach statistical significance, and there was no difference at the PP2 stage (day 11). This is likely because the pancreatic differentiation protocol has factors that drive cells toward a pancreatic identity, and virtually all cells already express PDX1 (Figures 3C and 3D). However, NKX6.1 expression was higher in iPSC−/− PP2 cells compared with iPSC+/+ PP2 cells. NKX6.1 is crucial for pancreatic progenitor identity and required for further differentiation to β cells (Rezania et al., 2013). In addition, expression of ISL1 was markedly increased, while MNX1 trended higher in iPSC−/− cells compared with iPSC+/+ cells (Figure 3D). In the ESC−/− line, MNX1 expression was significantly increased (Figure S3F). We also examined hepatic gene expression during the pancreatic differentiation. Levels of hepatic genes were lower in the iPSC−/− line compared with the iPSC+/+ line (Figure 3E), suggesting the loss of TBX3 generates a purer PP2 population. Similar results were seen in the ESC−/− line (Figures S3D–S3H). These data suggest that a lack of TBX3 improves differentiation efficiency and purity, as the iPSC−/− line generated more PP2 cells with increased expression of pancreas genes and decreased expression of hepatic genes, compared with the iPSC+/+ line.
Figure 3.
Loss of TBX3 enhances pancreatic progenitor generation from PSCs
(A) Schematic of directed differentiation protocol of PSCs to immature β cells.
(B) Representative example of PDX1 and NKX6.1 expression in PP2 cells by flow cytometry.
(C) Quantification of the percentage of PDX1+/NKX6.1+ and PDX1+ PP2 cells (n = 6 per cell line).
(D) Time course of PDX1, NKX6.1, ISL1, and MNX1 expression during pancreatic differentiation by qRT-PCR (n = 6 per time point, per cell line).
(E) Time course of AFP, TTR, ApoA2, and ApoB expression during pancreatic differentiation by qRT-PCR (n = 6 per time, point per cell line).
For all statistical analysis, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p ≤ 0.0001.
iPSC−/− cells are enriched for a pancreatic gene signature
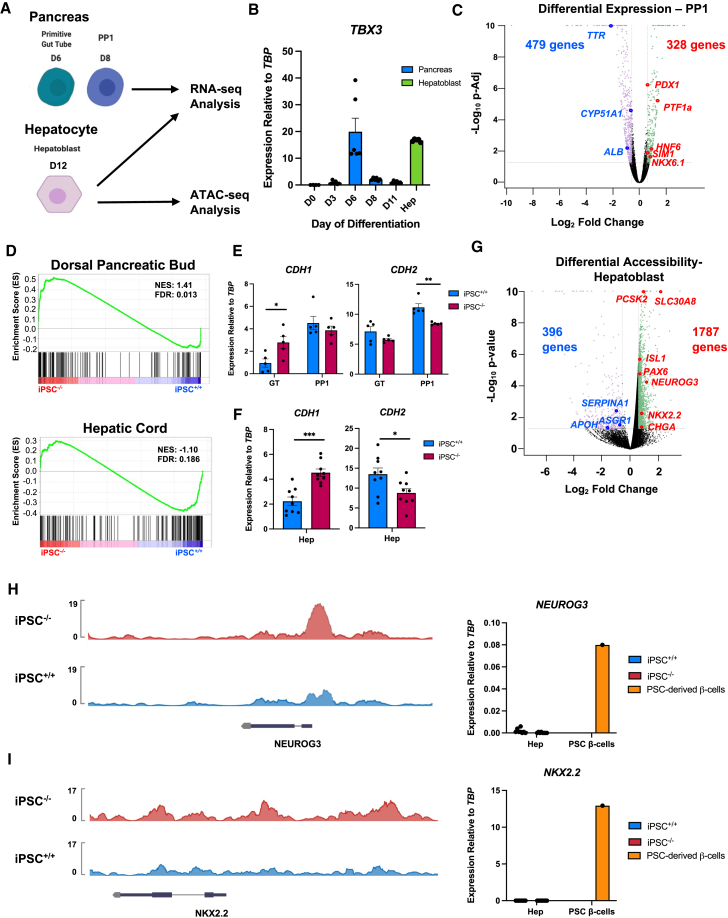
To investigate the effect of the loss of TBX3 on pancreatic differentiation in greater detail, genome-wide gene expression was examined using RNA sequencing (RNA-seq) in iPSC−/− versus iPSC+/+ lines at days 6 and 8 of pancreas differentiation (Figure 4A). We chose day 6 as TBX3 expression peaked at this point in the differentiation, and was comparable to TBX3 levels at day 12 during the hepatocyte differentiation (Figure 4B). This stage of differentiation is representative of the gut tube (GT) endoderm when endodermal cells are patterned to foregut. We chose day 8 PP1 cells, as this is when cells begin expressing PDX1. At both the GT and the PP1 stages, pancreatic genes were upregulated while hepatic genes were downregulated in iPSC−/− versus iPSC+/+ (Figures S4A and 4C). Pathway analysis showed that genes involved in metabolic pathways were downregulated in iPSC−/− GT cells compared with iPSC+/+ GT cells (Table S4A), and pathways relating to pancreatic development and function were upregulated in iPSC−/− PP1 (Table S4B). To determine if the gene expression signature of iPSC−/− cells in our in vitro differentiation was indicative of pancreas commitment at the expense of liver fate, we used gene set enrichment analysis with a previously published gene set from primary human embryos, comparing the dorsal pancreatic buds with hepatic cords (Jennings et al., 2017). Hepatic cord–specific genes were enriched in iPSC+/+ versus iPSC−/− GT cells, and pancreatic bud–specific genes were significantly enriched in the iPSC−/− PP1 population (Figures S4B and 4D). These data further confirm that the loss of TBX3 helped drive cells toward a pancreatic fate and away from a hepatic fate.
Figure 4.
iPSC−/− cells are enriched for a pancreatic gene signature
(A) Schematic of stages from PSC differentiations collected for sequencing analysis.
(B) Time course of TBX3 expression by qRT-PCR during pancreatic differentiation (n = 6) and hepatoblasts (n = 5) for comparison.
(C) Volcano plot of up- and downregulated genes in iPSC−/− versus iPSC+/+ pancreatic progenitor 1 (PP1) cells. p-Adj = 0.05, fold change: ≥ 1.5 and ≤ −1.5.
(D) Gene set enrichment analysis comparing normalized gene expression of samples in (C) to genes enriched in human fetal dorsal pancreatic bud and hepatic cord.
(E) Expression of CDH1 and CDH2 in GT and PP1 cells from pancreatic differentiation by qRT-PCR (n = 5 per time point, per cell line).
(F) Expression of CDH1 and CDH2 in hepatoblasts from hepatic differentiation by qRT-PCR (n = 9 per time point, per cell line).
(G) Volcano plot of differential accessibility of genes identified by ATAC-seq analysis in iPSC−/− versus iPSC+/+ hepatoblasts. p value = 0.05, fold change: ≥ 1.5 and ≤ −1.5.
(H) Representative example of tracks showing differential accessibility at the NEUROG3 and NKX2.2 loci. The y axis represents peak height.
(I) Expression of NEUROG3 and NKX2.2 in hepatoblasts (n = 8 per cell line) from hepatic differentiation and PSC-derived β cells (n = 1) by qRT-PCR.
For all statistical analysis, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p ≤ 0.0001.
Additional characterization was performed using RNA-seq analysis of iPSC−/– versus iPSC+/+ hepatoblasts (Figure 4A). PROX1, which is required for hepatoblast migration and is downregulated in Tbx3−/− mice (Lüdtke et al., 2009), and CDH2, encoding mesenchymal marker N-cadherin, were downregulated in iPSC−/− hepatoblasts, suggesting a migratory defect. Consistent with our prior gene expression findings, PDX1, ISL1, and MNX1 were upregulated in iPSC−/− hepatoblasts (Figures S4C). To better understand the global changes in gene expression due to the loss of TBX3, a covariate analysis was performed to identify commonly dysregulated genes in iPSC−/− cells at three developmental stages: GT and PP1 cells from the pancreatic differentiation, and hepatoblasts from the hepatocyte differentiation (Figures 4A and S4D). The covariate analysis showed 1,398 genes were similarly up- or downregulated in all three developmental stages (Figure S4E and Table S4C). Several pathways, including epithelial-mesenchymal transition (EMT), were dysregulated upon loss of TBX3, regardless of cell type (Table S4D), suggesting a common role of TBX3 in pancreas and liver development. At the GT and hepatoblast stages, CDH1, encoding epithelial marker E-cadherin, expression was higher in iPSC−/− cells than iPSC+/+ cells, while there was no significant difference at the PP1 stage. CDH2 expression was lower in iPSC−/− cells than iPSC+/+ cells at all three stages (Figures 4E and 4F). E-cadherin protein levels were higher in iPSC−/− cells than in iPSC+/+ cells, while the effect on N-cadherin protein was subtle (Figures S4F and S4G). Taken together, these data suggest that EMT may be dysregulated in liver and pancreas cells with the loss of TBX3. Although this dysregulation does not appear to have clear implications in impacting a liver versus pancreas fate, it may have roles at other developmental stages.
TBX3 promotes invasiveness of cancer cells through direct regulation of EMT genes SLUG (Krstic et al., 2019) and E-cadherin (Rodriguez et al., 2008). During liver bud expansion, hepatoblasts undergo EMT and delaminate into the surrounding mesenchyme. Tbx3−/− mice maintain E-cadherin expression, indicating a failure in EMT (Lüdtke et al., 2009). In pancreas development, endocrine progenitors undergo EMT and delaminate from the branching epithelium to form pancreatic islets (Gouzi et al., 2011). Although Tbx3 expression has been observed in the developing and adult mouse pancreas, its role is unclear. Based on the role of TBX3 in regulating EMT in other biological contexts, it may function similarly in delamination during islet formation. Cytoskeletal structure influences delamination (Kesavan et al., 2014) and differentiation (Mamidi et al., 2018) in the mouse, and enhances PP2 generation from PSCs (Hogrebe et al., 2020). We show that loss of TBX3 enhances PP2 generation, highlighting possible links among TBX3, cytoskeletal state, and differentiation.
A more detailed analysis of the ATAC-seq dataset from iPSC−/− versus iPSC+/+ hepatoblasts was performed and compared with the RNA-seq datasets; 1,787 genes had more accessible chromatin in iPSC−/− hepatoblasts, while 396 genes had less accessible chromatin (Figure 4G), highlighting the role of TBX3 as a transcriptional repressor. Upregulated genes from the RNA-seq covariate analysis largely overlap with genes identified by ATAC-seq as more open (Figures S4H and S4I). Pathway analysis of the ATAC-seq dataset showed that genes involved the same pathways such as EMT identified in the RNA-seq covariate analysis are differentially accessible, demonstrating good correlation between accessibility and gene expression (Table S4E). The top hit in the pathway analysis was upregulation of Hallmark Pancreas Beta Cells, confirming de-repression of the pancreatic program.
Interestingly, some genes involved in later endocrine cell development and maturation, such as NEUROG3 and NKX2.2, are more accessible in iPSC−/− than in iPSC+/+ hepatoblasts. Although they are more accessible, NEUROG3 and NKX2.2 are not expressed in iPSC−/− hepatoblasts (Figures 4H and 4I). These data suggest that TBX3 not only represses genes involved in early pancreatic development such as PDX1, but also is involved in keeping genes expressed later in pancreatic endocrine cell development in a less accessible state. Further studies on how the loss of TBX3 affects later pancreatic differentiation can enhance our understanding of endocrine cell development and improve our methodology for generating these cells from PSCs.
This study provides insight into the biology of patterning of the hepatic and pancreatic domains during human foregut development. We established that TBX3 is critical for proper hepatocyte development and maturation in humans, similar to the role of Tbx3 in mouse liver development. We also demonstrate that loss of TBX3 increases the number of pancreatic progenitors derived from PSCs. These results contribute to our knowledge of early endoderm patterning and provide a potential route to improve the generation of in vitro–derived pancreatic progenitors, which can be further differentiated into pancreatic cell types.
Experimental procedures
PSC lines
The CHOPi004-A iPSC line was made by the Human Pluripotent Stem Cell Core at the Children's Hospital of Philadelphia (Mukherjee et al., 2020). The Mel1 ESC line with a GFP reporter in the Insulin locus, and reverse tetracycline transactivator construct in the AAVS1 locus was used as a second genetic background (Micallef et al., 2012; Tiyaboonchai et al., 2017).
Genome editing using CRISPR/Cas9
TBX3 mutant PSC lines were generated, as described previously (Maguire et al., 2019), using two guide RNAs (gRNAs) to create a deletion in the endogenous TBX3 locus. TBX3 gRNA1, 5′-GGAGTGGATGAGCCTCTCC-3′, targeted the transcription site and TBX3 gRNA2, 5′-CAAAGGTAAACCATGTCAC-3′, targeted the boundary of exon 5 and intron 4 of the TBX3 locus. Single colonies were screened for the deletion by PCR and topoisomerase-based cloning, and sequenced for confirmation.
Hepatocyte differentiation
PSCs were differentiated to hepatocytes, as previously described (Ogawa et al., 2013), with modifications. PSCs were split on to 1:3 diluted Matrigel (Corning)-coated 6-well plates, and the differentiation was started when cells reached 80% to 90% confluency. Cells were differentiated until the hepatocyte maturation B stage (day 25). Detailed modifications can be found in the supplemental experimental procedures.
Pancreatic β-cell differentiation
PSCs were differentiated to the PP2 stage, as previously described (Rezania et al., 2014), with modifications. PSCs were split onto 1:30 Matrigel-coated 6-well plates, and differentiated as a monolayer until the end of stage 4 (day 11). Detailed modifications can be found in the supplemental experimental procedures.
Genomic analysis
For RNA-seq analysis, iPSC+/+ and iPSC−/− lines were harvested on day 12 of the hepatocyte differentiation protocol, and days 6 and 8 of the pancreatic differentiation protocol. Samples were collected from three biological replicates for each time point and for each genotype. RNA was extracted as described in the supplemental experimental procedures, and sent for sequencing. For ATAC-seq analysis, iPSC+/+ and iPSC−/− cells were harvested on day 12 of the hepatocyte differentiation protocol. Samples were collected from two biological replicates for each genotype and sent to Genewiz for tagmentation and sequencing. Data were analyzed by Rosalind (https://rosalind.onramp.bio/), with a HyperScale architecture developed by OnRamp BioInformatics, Inc. (San Diego, CA). A gene list comparing gene expression in dorsal pancreatic bud and hepatic cord tissues dissected from human embryos (Jennings et al., 2017) was used for the gene set enrichment analysis.
Data and code availability
The RNA-seq and ATAC-seq data reported in this study can be found using the GEO accession number GEO: GSE180528.
Statistical analysis
Results from multiple experiments are expressed as the mean ± SEM. An unpaired two-tailed Student's t test for groups with equal variance was performed to determine p values. All statistical analyses were performed on Prism version 8.4.3 for Mac (GraphPad Software). In the figures, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p ≤ 0.0001, and n denotes individual experiments.
Author contributions
SM designed and performed all experiments described in this study, interpreted data, and prepared the manuscript and figures. DLF and PG provided overall scientific guidance and manuscript preparation, review, and editing.
Conflicts of interest
The authors declare no competing interests.
Acknowledgments
All illustrations in the figures were created with BioRender.com. This study was supported by NIH grants R01 DK118155 and R01 DK123162.
Published: October 14, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2021.09.004.
Supplemental information
(A) Downregulated REACTOME pathways in iPSC−/− versus iPSC+/+ gut tube (GT) cells.
(B) Upregulated REACTOME pathways in iPSC−/− versus iPSC+/+ pancreatic progenitor 1 (PP1) cells.
(C) Up- and downregulated genes from stage-corrected covariate analysis in iPSC−/− versus iPSC+/+ of GT, PP1, and hepatoblast cells.
(D) Up- and downregulated MSigDB_HALLMARK pathways from covariate analysis.
(E) MSigDB_HALLMARK pathways with differentially accessible genes.
References
- Ahlgren U., Jonsson J., Edlund H. The morphogenesis of the pancreatic mesenchyme is uncoupled from that of the pancreatic epithelium in IPF1/PDX1-deficient mice. Development. 1996;122:1409–1416. doi: 10.1242/dev.122.5.1409. [DOI] [PubMed] [Google Scholar]
- Ahlgren U., Pfaff S.L., Jessell T.M., Edlund T., Edlund H. Independent requirement for ISL1 in formation of pancreatic mesenchyme and islet cells. Nature. 1997;385:257–260. doi: 10.1038/385257a0. [DOI] [PubMed] [Google Scholar]
- Bamshad M., Lin R.C., Law D.J., Watkins W.C., Krakowiak P.A., Moore M.E., Franceschini P., Lala R., Holmes L.B., Gebuhr T.C., et al. Mutations in human TBX3 alter limb, apocrine and genital development in ulnar-mammary syndrome. Nat. Genet. 1997;16:311–315. doi: 10.1038/ng0797-311. [DOI] [PubMed] [Google Scholar]
- Begum S., Papaioannou V.E. Dynamic expression of Tbx2 and Tbx3 in developing mouse pancreas. Gene Expr. Patterns. 2011;11:476–483. doi: 10.1016/j.gep.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson H., Ota S., Campbell C.E., Hurlin P.J. A dominant repression domain in Tbx3 mediates transcriptional repression and cell immortalization: relevance to mutations in Tbx3 that cause ulnar-mammary syndrome. Hum. Mol. Genet. 2001;10:2403–2413. doi: 10.1093/hmg/10.21.2403. [DOI] [PubMed] [Google Scholar]
- Claiborn K.C., Sachdeva M.M., Cannon C.E., Groff D.N., Singer J.D., Stoffers D.A. Pcif1 modulates Pdx1 protein stability and pancreatic β cell function and survival in mice. J. Clin. Invest. 2010;120:3713–3721. doi: 10.1172/JCI40440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amour K.A., Bang A.G., Eliazer S., Kelly O.G., Agulnick A.D., Smart N.G., Moorman M.A., Kroon E., Carpenter M.K., Baetge E.E. Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat. Biotechnol. 2006;24:1392–1401. doi: 10.1038/nbt1259. [DOI] [PubMed] [Google Scholar]
- Deutsch G., Jung J., Zheng M., Lóra J., Zaret K.S. A bipotential precursor population for pancreas and liver within the embryonic endoderm. Development. 2001;128:871–881. doi: 10.1242/dev.128.6.871. [DOI] [PubMed] [Google Scholar]
- Esmailpour T., Huang T. TBX3 promotes human embryonic stem cell proliferation and neuroepithelial differentiation in a differentiation stage-dependent manner. Stem Cells. 2012;30:2152–2163. doi: 10.1002/stem.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson-Brown J.J., Agulnik S.I., Silver L.M., Niswander L., Papaioannou V.E. Involvement of T-box genes Tbx2-Tbx5 in vertebrate limb specification and development. Development. 1998;125:2499–2509. doi: 10.1242/dev.125.13.2499. [DOI] [PubMed] [Google Scholar]
- Gouzi M., Kim Y.H., Katsumoto K., Johansson K., Grapin-Botton A. Neurogenin3 initiates stepwise delamination of differentiating endocrine cells during pancreas development. Dev. Dyn. 2011;240:589–604. doi: 10.1002/dvdy.22544. [DOI] [PubMed] [Google Scholar]
- Hebrok M., Kim S.K., Melton D.A. Notochord repression of endodermal sonic hedgehog permits pancreas development. Genes Dev. 1998;12:1705–1713. doi: 10.1101/gad.12.11.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogrebe N.J., Augsornworawat P., Maxwell K.G., Velazco-Cruz L., Millman J.R. Targeting the cytoskeleton to direct pancreatic differentiation of human pluripotent stem cells. Nat. Biotechnol. 2020;38:460–470. doi: 10.1038/s41587-020-0430-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings R.E., Berry A.A., Gerrard D.T., Wearne S.J., Strutt J., Withey S., Chhatriwala M., Piper Hanley K., Vallier L., Bobola N., et al. Laser capture and deep sequencing reveals the transcriptomic programmes regulating the onset of pancreas and liver differentiation in human embryos. Stem Cell Reports. 2017;9:1387–1394. doi: 10.1016/j.stemcr.2017.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J., Zheng M., Goldfarb M., Zaret K.S. Initiation of Mammalian liver development from endoderm by fibroblast Growth factors. Science. 1999;284:1998–2003. doi: 10.1126/science.284.5422.1998. [DOI] [PubMed] [Google Scholar]
- Kesavan G., Lieven O., Mamidi A., Öhlin Z.L., Johansson J.K., Li W.C., Lommel S., Greiner T.U., Semb H. Cdc42/N-WASP signaling links actin dynamics to pancreatic β cell delamination and differentiation. Development. 2014;141:685–696. doi: 10.1242/dev.100297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krstic M., Kolendowski B., Cecchini M.J., Postenka C.O., Hassan H.M., Andrews J., MacMillan C.D., Williams K.C., Leong H.S., Brackstone M., et al. TBX3 promotes progression of pre-invasive breast cancer cells by inducing EMT and directly up-regulating SLUG. J. Pathol. 2019;248:191–203. doi: 10.1002/path.5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Arber S., Jessell T.M., Edlund H. Selective agenesis of the dorsal pancreas in mice lacking homeobox gene Hlxb9. Nat. Genet. 1999;23:67–70. doi: 10.1038/12669. [DOI] [PubMed] [Google Scholar]
- Lüdtke T.H.W., Christoffels V.M., Petry M., Kispert A. Tbx3 promotes liver bud expansion during mouse development by suppression of cholangiocyte differentiation. Hepatology. 2009;49:969–978. doi: 10.1002/hep.22700. [DOI] [PubMed] [Google Scholar]
- Maguire J.A., Cardenas-Diaz F.L., Gadue P., French D.L. Highly efficient CRISPR-Cas9-mediated genome editing in human pluripotent stem cells. Curr. Protoc. Stem Cell Biol. 2019;48:1–14. doi: 10.1002/cpsc.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamidi A., Prawiro C., Seymour P.A., de Lichtenberg K.H., Jackson A., Serup P., Semb H. Mechanosignalling via integrins directs fate decisions of pancreatic progenitors. Nature. 2018;564:114–118. doi: 10.1038/s41586-018-0762-2. [DOI] [PubMed] [Google Scholar]
- Micallef S.J., Li X., Schiesser J.V., Hirst C.E., Yu Q.C., Lim S.M., Nostro M.C., Elliott D.A., Sarangi F., Harrison L.C., et al. INSGFP/w human embryonic stem cells facilitate isolation of in vitro derived insulin-producing cells. Diabetologia. 2012;55:694–706. doi: 10.1007/s00125-011-2379-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Gagne A.L., Maguire J.A., Jobaliya C.D., Mills J.A., Gadue P., French D.L. Generation of human control iPSC line CHOPi004-A from juvenile foreskin fibroblast cells. Stem Cell Res. 2020;49:102084. doi: 10.1016/j.scr.2020.102084. [DOI] [PubMed] [Google Scholar]
- Nostro M.C., Sarangi F., Ogawa S., Holtzinger A., Corneo B., Li X., Micallef S.J., Park I.-H., Basford C., Wheeler M.B., et al. Stage-specific signaling through TGF family members and WNT regulates patterning and pancreatic specification of human pluripotent stem cells. Development. 2011;138:1445. doi: 10.1242/dev.055236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa S., Surapisitchat J., Virtanen C., Ogawa M., Niapour M., Sugamori K.S., Wang S., Tamblyn L., Guillemette C., Hoffmann E., et al. Three-dimensional culture and cAMP signaling promote the maturation of human pluripotent stem cell-derived hepatocytes. Development. 2013;140:3285–3296. doi: 10.1242/dev.090266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezania A., Bruin J.E., Xu J., Narayan K., Fox J.K., O’Neil J.J., Kieffer T.J. Enrichment of human embryonic stem cell-derived NKX6.1-Expressing pancreatic progenitor cells accelerates the maturation of insulin-secreting cells in vivo. Stem Cells. 2013;31:2432–2442. doi: 10.1002/stem.1489. [DOI] [PubMed] [Google Scholar]
- Rezania A., Bruin J.E., Arora P., Rubin A., Batushansky I., Asadi A., O’Dwyer S., Quiskamp N., Mojibian M., Albrecht T., et al. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat. Biotechnol. 2014;32:1121–1133. doi: 10.1038/nbt.3033. [DOI] [PubMed] [Google Scholar]
- Rodriguez M., Aladowicz E., Lanfrancone L., Goding C.R. Tbx3 represses E-cadherin expression and enhances melanoma invasiveness. Cancer Res. 2008;68:7872–7881. doi: 10.1158/0008-5472.CAN-08-0301. [DOI] [PubMed] [Google Scholar]
- Rossi J.M., Dunn N.R., Hogan B.L.M., Zaret K.S. Distinct mesodermal signals, including BMPs from the septum, transversum mesenchyme, are required in combination for hepatogenesis from the endoderm. Genes Dev. 2001;15:1998–2009. doi: 10.1101/gad.904601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si-Tayeb K., Noto F.K., Nagaoka M., Li J., Battle M.A., Duris C., North P.E., Dalton S., Duncan S.A. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology. 2010;51:297–305. doi: 10.1002/hep.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Hoogaars W.M., Barnett P., Grieskamp T., Sameer Rana M., Buermans H., Farin H.F., Petry M., Heallen T., Martin J.F., et al. Tbx2 and Tbx3 induce atrioventricular myocardial development and endocardial cushion formation. Cell. Mol. Life Sci. 2012;69:1377–1389. doi: 10.1007/s00018-011-0884-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo A.K.K., Tsuneyoshi N., Hoon S., Tan E.K., Stanton L.W., Wright C.V.E., Dunn N.R. PDX1 binds and represes hepatic genes to ensure robust pancreatic commitment in differentiating human embryonic stem cells. Stem Cell Reports. 2015;4:578–590. doi: 10.1016/j.stemcr.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiyaboonchai A., Cardenas-Diaz F.L., Ying L., Maguire J.A., Sim X., Jobaliya C., Gagne A.L., Kishore S., Stanescu D.E., Hughes N., et al. GATA6 plays an important role in the induction of human definitive endoderm, development of the pancreas, and functionality of pancreatic β cells. Stem Cell Reports. 2017;8:589–604. doi: 10.1016/j.stemcr.2016.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidgang C.E., Russell R., Tata P.R., Kühl S.J., Illing A., Müller M., Lin Q., Brunner C., Boeckers T.M., Bauer K., et al. TBX3 directs cell-fate decision toward mesendoderm. Stem Cell Reports. 2013;1:248–265. doi: 10.1016/j.stemcr.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorn A.M., Wells J.M. Vertebrate endoderm development and organ formation. Annu. Rev. Cell Dev. Biol. 2009;25:221–251. doi: 10.1146/annurev.cellbio.042308.113344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Downregulated REACTOME pathways in iPSC−/− versus iPSC+/+ gut tube (GT) cells.
(B) Upregulated REACTOME pathways in iPSC−/− versus iPSC+/+ pancreatic progenitor 1 (PP1) cells.
(C) Up- and downregulated genes from stage-corrected covariate analysis in iPSC−/− versus iPSC+/+ of GT, PP1, and hepatoblast cells.
(D) Up- and downregulated MSigDB_HALLMARK pathways from covariate analysis.
(E) MSigDB_HALLMARK pathways with differentially accessible genes.
Data Availability Statement
The RNA-seq and ATAC-seq data reported in this study can be found using the GEO accession number GEO: GSE180528.