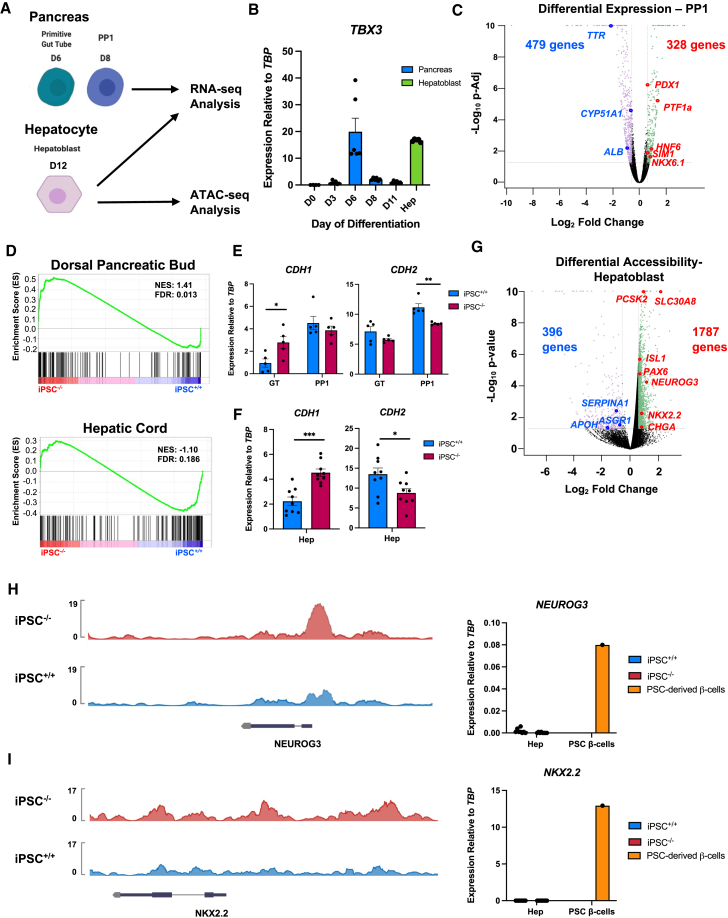
Figure 4.
iPSC−/− cells are enriched for a pancreatic gene signature
(A) Schematic of stages from PSC differentiations collected for sequencing analysis.
(B) Time course of TBX3 expression by qRT-PCR during pancreatic differentiation (n = 6) and hepatoblasts (n = 5) for comparison.
(C) Volcano plot of up- and downregulated genes in iPSC−/− versus iPSC+/+ pancreatic progenitor 1 (PP1) cells. p-Adj = 0.05, fold change: ≥ 1.5 and ≤ −1.5.
(D) Gene set enrichment analysis comparing normalized gene expression of samples in (C) to genes enriched in human fetal dorsal pancreatic bud and hepatic cord.
(E) Expression of CDH1 and CDH2 in GT and PP1 cells from pancreatic differentiation by qRT-PCR (n = 5 per time point, per cell line).
(F) Expression of CDH1 and CDH2 in hepatoblasts from hepatic differentiation by qRT-PCR (n = 9 per time point, per cell line).
(G) Volcano plot of differential accessibility of genes identified by ATAC-seq analysis in iPSC−/− versus iPSC+/+ hepatoblasts. p value = 0.05, fold change: ≥ 1.5 and ≤ −1.5.
(H) Representative example of tracks showing differential accessibility at the NEUROG3 and NKX2.2 loci. The y axis represents peak height.
(I) Expression of NEUROG3 and NKX2.2 in hepatoblasts (n = 8 per cell line) from hepatic differentiation and PSC-derived β cells (n = 1) by qRT-PCR.
For all statistical analysis, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p ≤ 0.0001.