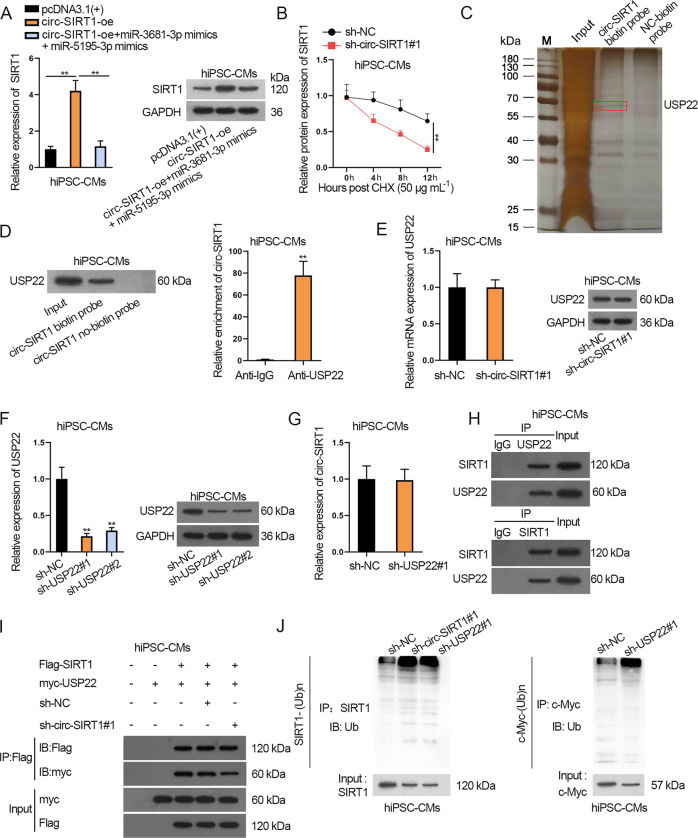
Fig. 5. Circ-SIRT1 recruits USP22 to regulate the deubiquitination and stabilization of SIRT1 in hiPSC-CMs.
A SIRT1 mRNA and protein expression in hiPSC-CMs transfected with pcDNA3.1(+), circ-SIRT1-oe, or circ-SIRT1-oe+miR-3681-3p mimics+miR-5195-3p mimics were analyzed via RT-qPCR and western blot analyses. N = 3. B SIRT1 protein stability was analyzed by western blot at 0, 4, 8, and 12 h after CHX treatment in hiPSC-CMs. N = 3. C Silver staining of differential proteins in RNA pull-down products for circ-SIRT1 biotin probe compared with NC-biotin probe. The differential bands were sent to mass spectrometry and USP22 was shown to be significantly enriched in circ-SIRT1 biotin probe group in hiPSC-CMs. N = 3. D Western blot band of USP22 in RNA pull-down products for circ-SIRT1 biotin probe and circ-SIRT1 no-biotin probe. RT-qPCR results of circ-SIRT1 enrichment in RIP precipitates for anti-IgG and anti-USP22. N = 3. E RT-qPCR and western blot analyzed the effect of silenced circ-SIRT1 on USP22 expression. N = 3. F RT-qPCR and western blot analyses of the efficiency of USP22 knockdown by sh-USP22#1/2 in hiPSC-CMs. N = 3. G RT-qPCR analysis of the effect of silenced USP22 on circ-SIRT1 expression. N = 3. H Western blot band of SIRT1 and USP22 in the CoIP products for anti-IgG and anti-USP22. N = 3. I The hiPSC-CMs were transfected with exogenous Flag-tagged SIRT1 and myc-tagged USP22 proteins and sh-NC or sh-circ-SIRT1#1. Western blot band showed the enrichment of myc in the CoIP products in anti-Flag in sh-NC and sh-circ-SIRT1#1 groups. N = 3. J Analysis of the effect of circ-SIRT1 depletion or USP22 deficiency on SIRT1 ubiquitination is shown, taking c-Myc as a positive control for IP. N = 3. **P < 0.01 was assessed by Student’s t test for comparison between groups, one-way ANOVA and Tukey for multiple groups.