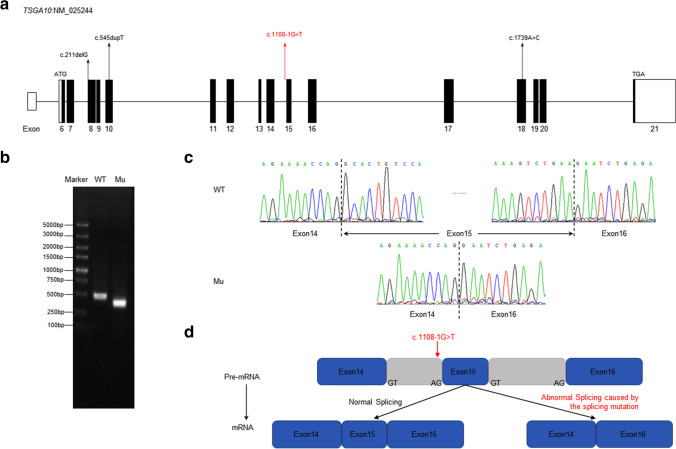
Fig. 3.
The location and conservation of the mutant residue in TSGA10. a The location of mutations in the TSGA10 exons. Mutations described in the previous study are marked in black and the novel splicing mutation identified in our study is marked in red. b Agarose gel electrophoresis illustrating the effect of the TSGA10 c.1108-1G > T mutation. Compared with the wild type lane, the mutation lane showed a smaller band. c Identified the abnormal splicing isoform of the mutation. By RT-PCR and Sanger sequencing, the results showed that the splice-site mutation in TSGA10 lead to the skipping of exon15. d A possible scheme illustrating of the effect of the TSGA10 c.1108-1G > T mutation. In this case, the mutation skips the canonical receptor site leading to the entire deletion of exon15 in cDNA and resulting in abnormal transcription and translation