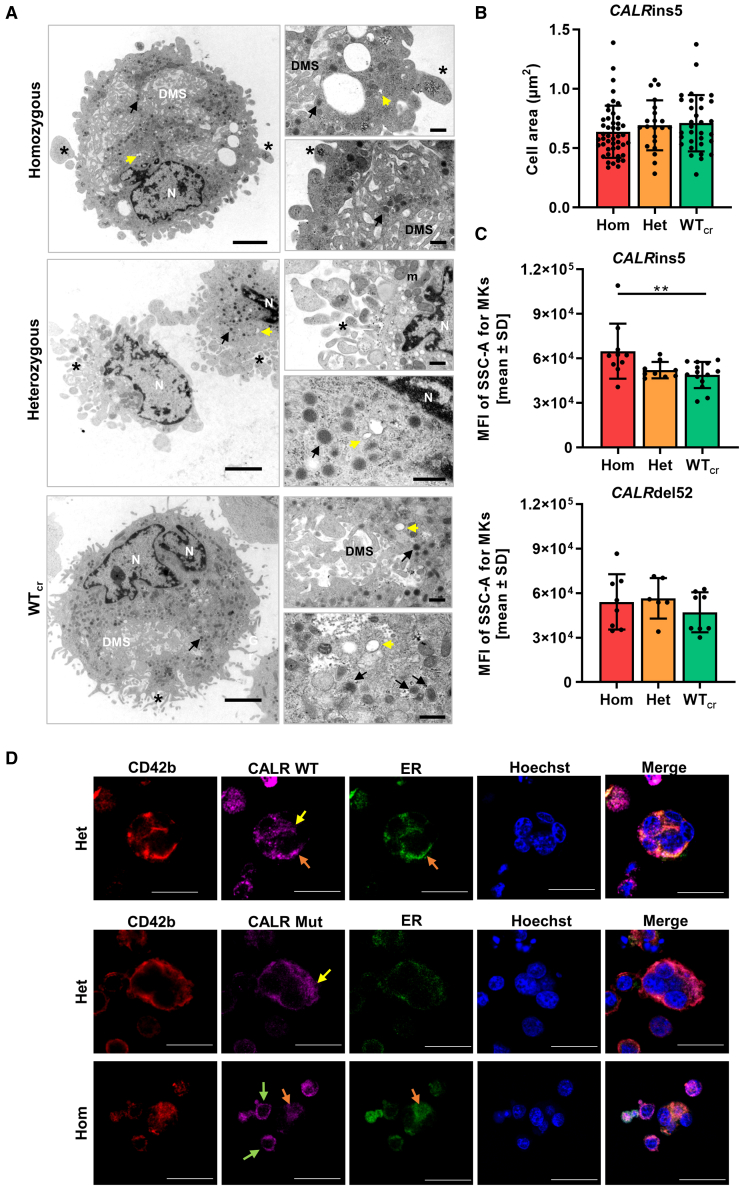
Figure 6.
Structural morphology analysis of iPSC-derived MKs and CALR distribution in MKs
(A) Transmission electron microscopy (TEM) images of iPSC-derived MKs of indicated CALRins5 genotype to evaluate cell morphology. Representative images are shown. N, nucleus; DMS, demarcation membrane system; m, mitochondria. Asterisks indicate pro-platelet protrusions. Black arrows point to granules and yellow arrows indicate open canalicular system. Scale bars, 2.5 μm (large panels) and 500 nm (small panels).
(B) Calculated cell area of MKs in TEM images for indicated CALRins5 genotypes. Each data point represents a single cell, n = 1 differentiation experiment. Data are presented as mean ± SD.
(C) Mean fluorescence intensity (MFI) calculated for the side scatter (SSC-A) of CD42b+CD41+CD61+ MKs on day 14 of “spin-EB” differentiation. Indicated data points represent independent experiments for each CALRins5 (n = 10–14) and CALRdel52 (n = 6–8) genotype. Values are shown as mean ± SD; ∗∗p < 0.01.
(D) Representative immunofluorescence images of iPSC-derived MKs stained for the ER and WT CALR or mutated (Mut) CALR after 14 days of differentiation for indicated CALRins5 iPSC clones. To identify MKs, samples were additionally stained for CD42b. Hoechst was added for nuclear staining. Diffuse CALR distribution, clustered localization of CALR at the cell surface, and co-localization of CALR and ER are indicated by yellow, green, and orange arrows, respectively. Scale bars, 50 μm.