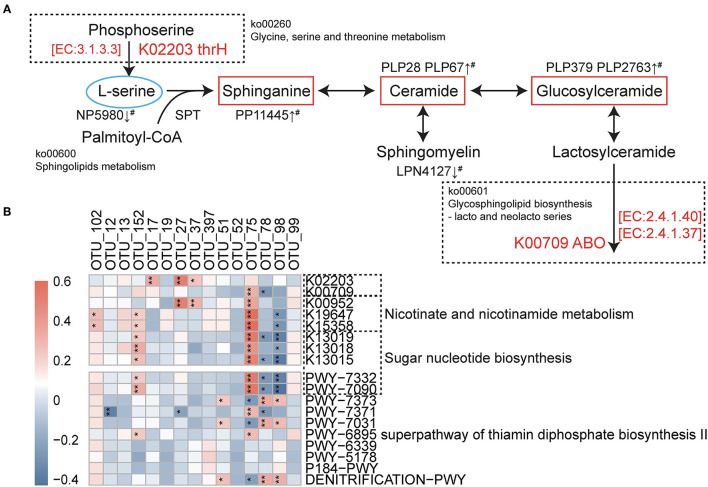
Figure 4.
Association between predicted metagenomic functional changes and differential OTUs as well as sphingolipids metabolism. (A) Relationship between sphingolipids pathway and two related differential enzymes (K02203 and K00709) predicted by PICRUSt2. The features are highlighted in red (smoking-positive) or blue (smoking-negative). #Observed metabolomic changes in smokers with CAD compared with never smokers in the present study. IDs and names of enzymes and pathways were based on the KEGG database. SPT, serine pamitoyl-transferase; K02203, homoserine phosphotransferase; K00709, histo-blood group ABO system transferase. (B) Spearman correlations between smoking-associated OTUs and metagenomic prediction including differential pathways as well as important enzymes. IDs of enzymes were according to the KEGG database; IDs of pathways were according to the MetaCyc database. *p < 0.05, **p < 0.01.