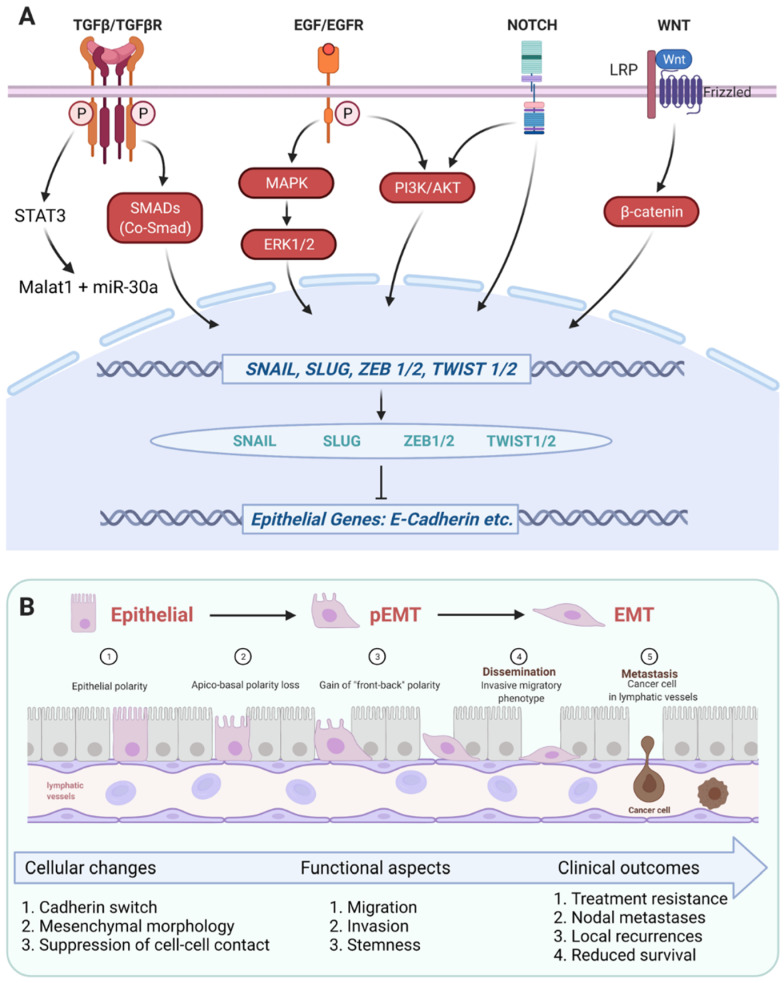
Figure 2.
Schematic representation of central signaling pathways involved in the induction of (partial) EMT in HNSCC. (A) Figure 2A depicts major ligands, receptors, signaling components (red color) and transcription factors involved in the induction of EMT and partial EMT (genes are depicted in blue color, proteins in green color) via the repression of epithelial genes (depicted in blue color). TGFβR, EGFR, NOTCH, and LRP/Frizzled receptors have been reported to induce the expression of EMT-TFs SNAIL, SLUG, ZEB1/2, and TWIST1/2 through the binding of their cognate ligands, i.e., TGFβ, EGF, Delta/Jagged or mutations, and WNT variants, respectively. TGFβR signaling towards EMT is functional via signal transducer and activator of transcription 3 (STAT3) and the subsequent induction of Malat1 and miR-30a, and via SMADs/co-SMAD. Activation of EMT through EGF/EGFR was reported to depend primarily on MAPK and ERK1/2; however, induction via PI3K and AKT was described too. Uncleavable mutated variants of NOTCH receptors were shown to trans-activate EGFR signaling at the level of PI3K and AKT, converging in EMT induction. WNT signaling via the Frizzled/LRP receptors result in nuclear translocation of β-catenin. (B) Figure 2B depicts the cellular, functional, and clinical consequences of the induction of (p)EMT described in Figure 2A. Molecular and cellular changes associated with (p)EMT include a progressive transit from an epithelial polarity (①) to a loss of apico-basal polarity (②) and a gain of “front-back polarity” (③), a switch in cadherin variants expression from E-cadherin to N-cadherin, the adoption of a mesenchymal morphology due to a suppression of cell–cell contacts. As a result, cells in EMT are characterized by enhanced migration, invasion, and stem-like properties (④) that influence treatment resistance, metastases formation (⑤), and recurrences, and thereby affect the clinical outcome of HNSCC patients (Figure generated using BioRender, Toronto, ON, Canada).