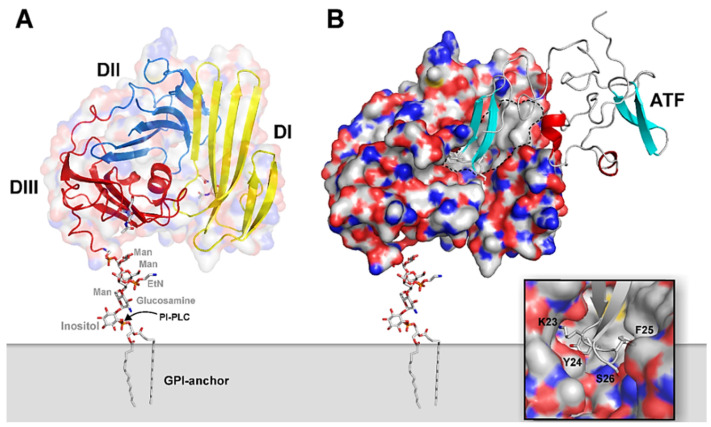
Figure 1.
Graphical representation of the structure of uPA•uPAR complexes on the cell surface. A composite molecular model representing human uPAR based on the crystal structure solved for uPAR•ATF complexes is displayed in (A) using the PDB coordinates 2FD6 [36]. The molecular shape of uPAR is visualized by a semitransparent surface, while secondary structure elements are depicted as ribbons. The assembly of the three LU-domains is evident from the color coding, yellow (DI), blue (DII), and red (DIII). A hypothetical model for the GPI-anchor, tethering uPAR to the cell surface, is shown as sticks. In (B), the bimolecular complex of uPAR with its natural binding ligand uPA is illustrated using a solid surface representation for uPAR and a ribbon diagram for the receptor-binding fragment of uPA (ATF) used to crystallize the complex. The large hydrophobic ligand-binding cavity of uPAR is highlighted by the grey area delimited by the hatched black line using the following atomic color coding: grey (C), blue (N), red (O), and yellow (S). The inset in the bottom right corner provides a more detailed illustration of the tight engagement and burial of the tip of the β-hairpin of GFD in uPA within the deepest region of the central cavity in uPAR. Adapted from [23].