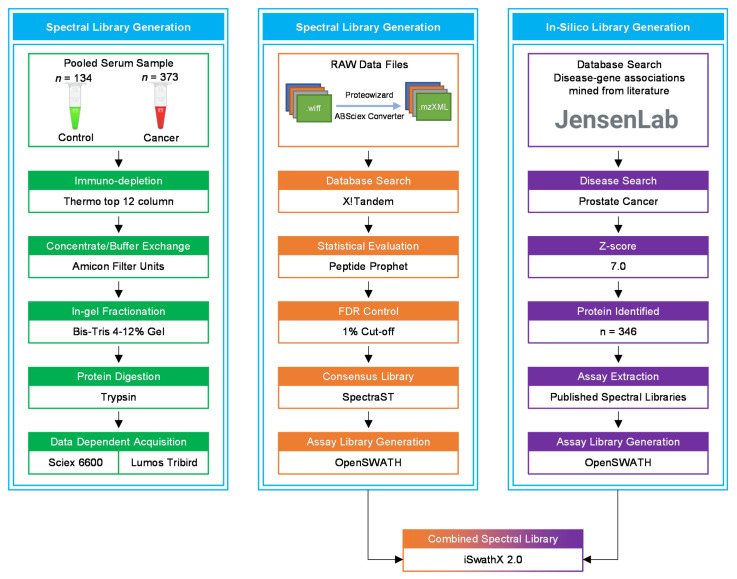
Figure 8.
A flowchart of the data acquisition and spectral library generation workflow. The pooled samples were immuno-depleted of the top 12 abundant proteins, followed by protein concentration, in-gel fractionation, and in-gel tryptic digestion. All samples were analysed using LC-MS/MS for data dependent acquisition (DDA). In the OpenSWATH library generation pipeline, X!Tandem was utilised for database search of raw MS files. The in silico library was created by extracting PCa-related proteins from the Disease database using a z-score cut-off of 5.0. iSwathX, a web-based tool, was used to merge all four assay libraries.