Figure 4.
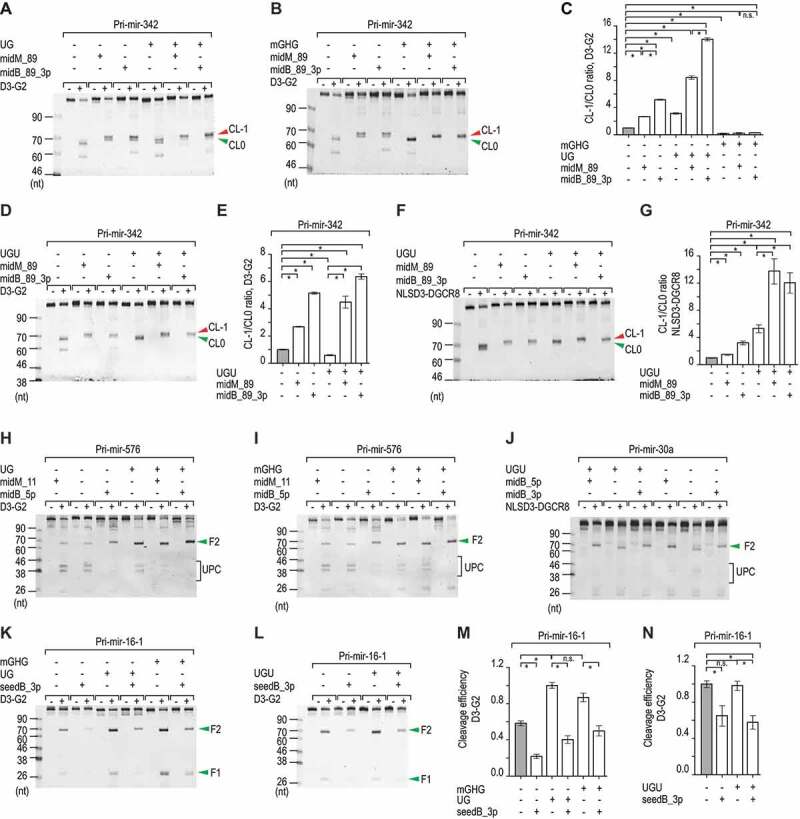
Coordination of bulges and known motifs. (A, B, D, F, H–L) Representative gel images showing the in vitro processing of the pri-mir-342, pri-mir-576, pri-mir-30a, and pri-mir-16-1 variants by D3-G2 or NLSD3-DGCR8. A 5 pmol aliquot of each pri-miRNA variant was incubated with 4–20 pmol D3-G2 or 4–8 pmol NLSD3-DGCR8 at 37°C for 120 min. UPC: unproductive cleavage products. (C, E and G) Bar charts showing the mean +/− SEM CL-1/CL0 ratios estimated from three repeated experiments for the pri-mir-342 variants. The band densities of the F2 fragments resulting from the CL-1 or CL0 cleavages for each variant were quantified by Image Lab v.6.0.1, and the CL-1/CL0 ratio of each variant was normalized to that of pri-mir-342 (midM_89). The asterisks (*) and (n.s.) indicate statistically significant (p < 0.05) and no significant differences, respectively, when the data were compared using the two-sided t-test. For (C), none versus midB_89: p = 9.6e-7, none versus UG+none: p = 6.4e-5, midB_89_3p versus UG+midB_89: p = 2.8e-6, none versus mGHG+none: p = 4.9e-4, midB_89_3p versus mGHG+midB_89_3p: p = 5.0e-7. For (E), none versus UGU+none: p = 0.002, midB_89_3p versus UGU+midB_89_3p: p = 0.004. For (G), none versus UGU+none: p = 0.002, midB_89_3p versus UGU+midB_89_3p: p = 0.005. (M and N) Bar charts to show the cleavage efficiency of Microprocessor, which was estimated from the three repeated processing experiments conducted for the pri-mir-16-1 variants. The band densities of the F2 fragments or the original substrates were quantified by Image Lab v.6.0.1. The cleavage efficiency was estimated as the ratio of the F2/original substrate. The ratio was normalized to that of the ‘UG’ variant (M) or to the ‘none’ variant (N). Data are presented as mean +/− SEM, and the asterisk (*) and (n.s.) indicate statistically significant (p < 0.05) and no significant differences, respectively when the data were compared using the two-sided t-test. For (M), none versus seedB_3p: p = 4.7e-4, UG+none versus UG+seedB_3p: p = 3.5e-4, and mGHG+none versus mGHG+seedB_3p: p = 0.007. For (N), none versus seedB_3p: p = 0.04, and UGU+none versus UGU+seedB_3p: p = 0.012
