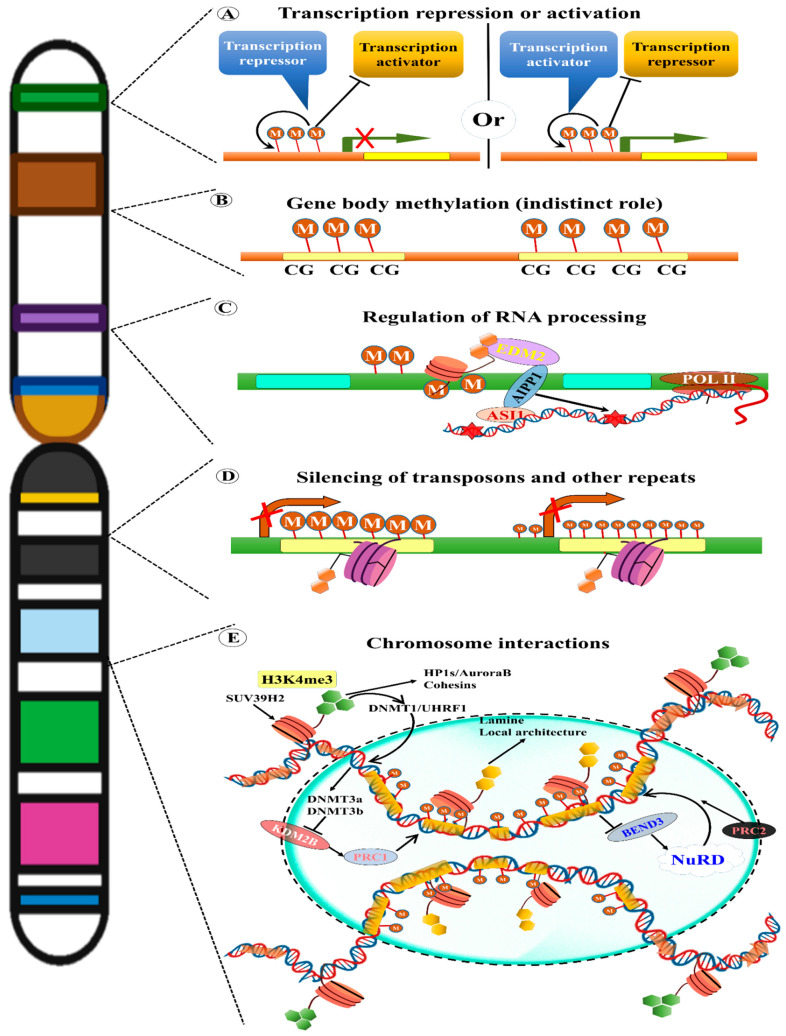
Figure 2.
Cellular functions of DNA methylation (m) in the plant genome. DNA methylation regulates transposon activation, gene regulation, and chromosome interactions. (A) Methylation in the gene promoter either represses or activates transcription [229,230,231,232,233]. (B) Gene body methylations mainly occur in the CG context, although its function remains unknown [42,231,234,235,236]. (C) DNA methylation in heterochromatin regions causes the ASI1-AIPP1-EDM2 complex to enhance polyadenylation sites (red stars). ASI1 binds RNA and associates with chromatin, and EDM2 catches demethylated histone H3 lysine in the heterochromatin region [159,237,238,239]. (D) The methylation of transposons and other DNA repeats mainly occurs in pericentromeric heterochromatin regions [231,235]. (E) Chromosome interactions among pericentromeric and heterochromatin islands are regulated by DNA methylation, and repressive chromatin regions are characterized by abundant transposons and small RNAs [240,241]. ASI1, anti-silencing 1; AIPP1, immunoprecipitated protein 1; EDM2, enhanced downy mildew 2; POL II, RNA polymerase II. The illustration was adapted and redrawn from Zhang et al. [42], with copyright permission from the Licensor Springer Nature (Nature Reviews Molecular Cell Biology: Nature publisher) and Copyright Clearance Center (https://www.copyright.com) (Supplementary File S4).