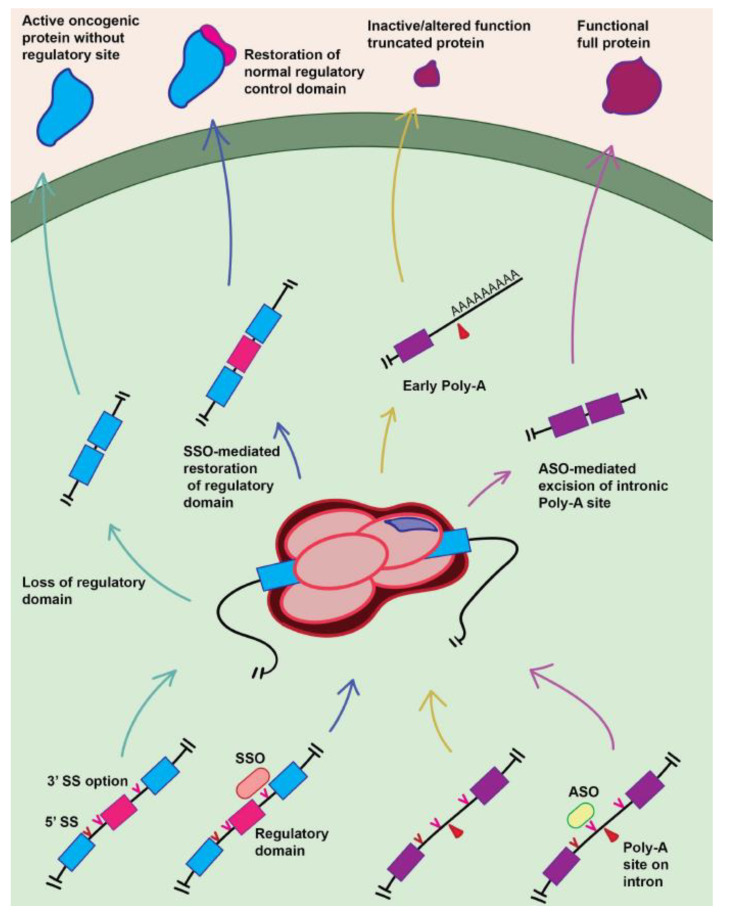
Figure 3.
Oligonucleotide-based therapeutic modulation of splice site selectivity by endogenous splicing machinery. The presence of multiple different splice site options that all produce a viable mRNA transcript post-processing opens the possibility of incorrect selection of the correct splice site for the tissue or cell, with potential tumorigenic effects. SSOs can be used to bind splice sites and prevent recognition, therefore allowing the modulation of the produced protein. For instance, the figure shows a case where dysregulated splicing pathways cause incorrect splice site recognition, leading to the production of an oncogenic protein lacking a regulatory site (and thus presumably being constitutively active, promoting uncontrolled growth). The introduction of SSOs restores the proper splicing product, producing a normally controlled protein. More generally, ASOs can be used for a similar function. The figure depicts a case where dysfunctional splicing machinery leads to improper inclusion of an intron which contains within it a polyadenylation site, leading to premature polyadenylation and truncation of the product. Introduction of the ASO blocks recognition of this splice site that was allowing for the inclusion of the intronic polyadenylation site, restoring the normal protein product. Abbreviations: ASO, antisense oligonucleotide; poly-A, polyadenylation; SS, splice site; SSO, splice-switching antisense oligonucleotide.