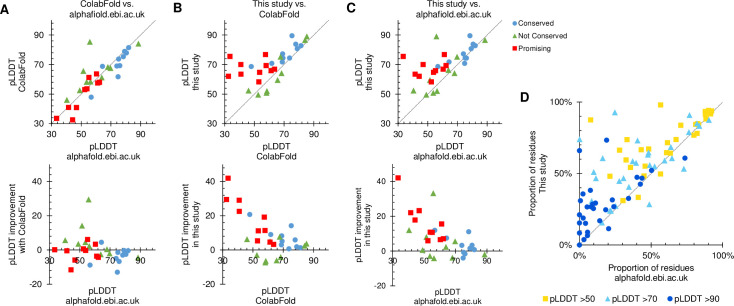
Fig 2. Improved AlphaFold2 predictions using ColabFold and a wider set of Discoba sequences for MSAs.
A-C) Comparison of per-protein average pLDDT for 30 test proteins, 10 random widely conserved proteins, 10 random kinetoplastids-specific proteins and 10 ‘promising’ kinetoplastid-specific proteins which appeared likely to improve with additional MSA sequences. A) Unmodified ColabFold in comparison to the AlphaFold database plotted as: Top, raw pLDDTs. Points to the top left of the diagonal represent improved (higher pLDDT) predictions. Bottom, change in pLDDT. Points above the horizontal axis represent improvement. B) This study (ColabFold with HMMER search of additional Discoba sequences) in comparison to unmodified ColabFold. C) This study in comparison to the AlphaFold database. D) The same comparison as C) but plotting the proportion of residues over different threshold pLDDT values instead of mean pLDDT.