Fig. 5.
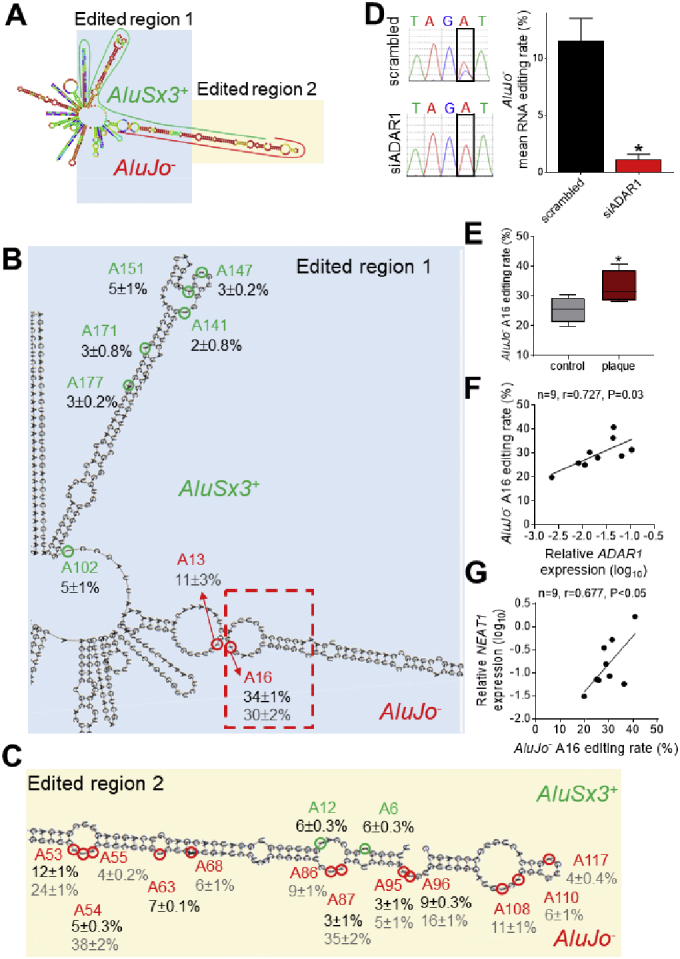
RNA editing mapping of NEAT1 lncRNA. A. RNA foldback structure of NEAT1 lncRNA region containing AluSx3+, MIR+ and AluJo‐ after editing of adenosine residues validated by the in-house PCR direct method. B,C. Mapping of A-to-I RNA editing sites within NEAT1 AluSx3+ and AluJo−. Numbers in black represent mean ± SEM of the editing rate detected in 5 independent biological replicates of cultured HUVECs. Numbers in grey represent the mean ± SEM in the 9 arterial tissue samples. D, left panel. Example of Sanger-sequencing chromatopherogram representing an A-to-G substitution of AluJo−; right panel, Quantification of average RNA editing rate in the adenosine residues of NEAT1 AluJo− in scrambled vs siADAR1 transfected HUVECs. Bars represent mean + SEM of n = 5 individual biological replicates per condition. E. Average editing rate of NEAT1 AluJo−A16 in human atherosclerotic plaques derived from carotid endarterectomy (n = 5) vs healthy arterial tissue (n = 4). F,G. Scatter-plots depicting the relationship between editing rate of NEAT1 AluJo−A16 and ADAR1 (F) or NEAT1 (G) expression in human arterial tissue. Correlation co-efficient derived by Pearson test. *P < 0.05.
