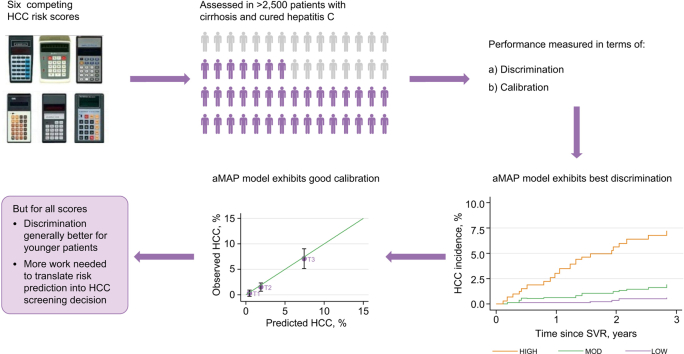
Abstract
Background & Aims
Hepatocellular carcinoma (HCC) prediction models can inform clinical decisions about HCC screening provided their predictions are robust. We conducted an external validation of 6 HCC prediction models for UK patients with cirrhosis and a HCV virological cure.
Methods
Patients with cirrhosis and cured HCV were identified from the Scotland HCV clinical database (N = 2,139) and the STratified medicine to Optimise Treatment of Hepatitis C Virus (STOP-HCV) study (N = 606). We calculated patient values for 4 competing non-genetic HCC prediction models, plus 2 genetic models (for the STOP-HCV cohort only). Follow-up began at the date of sustained virological response (SVR) achievement. HCC diagnoses were identified through linkage to nation-wide cancer, hospitalisation, and mortality registries. We compared discrimination and calibration measures between prediction models.
Results
Mean follow-up was 3.4–3.9 years, with 118 (Scotland) and 40 (STOP-HCV) incident HCCs observed. The age-male sex-ALBI-platelet count score (aMAP) model showed the best discrimination; for example, the Concordance index (C-index) in the Scottish cohort was 0.77 (95% CI 0.73–0.81). However, for all models, discrimination varied by cohort (being better for the Scottish cohort) and by age (being better for younger patients). In addition, genetic models performed better in patients with HCV genotype 3. The observed 3-year HCC risk was 3.3% (95% CI 2.6–4.2) and 5.1% (3.5–7.0%) in the Scottish and STOP-HCV cohorts, respectively. These were most closely matched by aMAP, in which the mean predicted 3-year risk was 3.6% and 5.0% in the Scottish and STOP-HCV cohorts, respectively.
Conclusions
aMAP was the best-performing model in terms of both discrimination and calibration and, therefore, should be used as a benchmark for rival models to surpass. This study underlines the opportunity for ‘real-world’ risk stratification in patients with cirrhosis and cured HCV. However, auxiliary research is needed to help translate an HCC risk prediction into an HCC-screening decision.
Lay summary
Patients with cirrhosis and cured HCV are at high risk of developing liver cancer, although the risk varies substantially from one patient to the next. Risk calculator tools can alert clinicians to patients at high risk and thereby influence decision-making. In this study, we tested the performance of 6 risk calculators in more than 2,500 patients with cirrhosis and cured HCV. We show that some risk calculators are considerably better than others. Overall, we found that the ‘aMAP’ calculator worked the best, but more work is needed to convert predictions into clinical decisions.
Keywords: Prognosis, Risk prediction, Primary liver cancer, External validation, Genetic risk scores
Abbreviations: aMAP, age-male sex-ALBI-platelet count score; ALT, alanine aminotransferase; AST, aspartate aminotransferase; C-index, Concordance index; GGT, gamma glutamyl transferase; GRS, genetic risk score; HCC, hepatocellular carcinoma; ICD, International Classification of Diseases; IDU, injecting-drug user; IF, interferon; PNPLA3, patatin-like phospholipase domain-containing protein 3; SMR01, Scottish Inpatient Hospital Admission Database; SMR06, Scottish Cancer Register; STOP-HCV, STratified medicine to Optimise Treatment of Hepatitis C Virus; SVR, sustained virological response; THRI, Toronto HCC Risk Index; VHA, Veteran Health Affairs
Graphical abstract
Highlights
-
•
The aMAP HCC model exhibits the best discrimination and calibration in patients with cirrhosis and SVR compared with competing models.
-
•
In general, the discriminative ability of non-genetic models declines with age.
-
•
Discriminative ability of genetic risk scores can vary by HCV genotype.
-
•
Some models underpredict HCC risk (i.e. underestimate the 3-year HCC probability).
-
•
Supporting research is needed to translate an HCC risk prediction into an HCC-screening decision.
Introduction
Patients with HCV-related cirrhosis remain at high risk of developing hepatocellular carcinoma (HCC) after a virological cure,[1], [2], [3] which does not appear to diminish over time.4 HCC has among the worst 5-year survival probabilities of any cancer.5 However, if detected at an early stage (i.e. when curative treatments can be administered), 5-year survival can exceed 70%.5 The current standard-of-care-for early HCC detection is biannual abdominal ultrasound surveillance with or without alpha foetoprotein.6,7 Although existing clinical guidelines recommend this intervention for all patients with cirrhosis after HCV eradication, there is growing recognition that a more targeted approach is needed (i.e. in which clinicians focus their finite resources on patients who stand to benefit the most from surveillance).8,9
It is against this backdrop that HCC prediction models are now emerging that can estimate a patient’s risk of developing HCC from routine data. Currently, such scores include the age-male sex-ALBI-platelet count score (aMAP),10 the Toronto HCC risk Index (THRI),11 models derived from the US Veteran Health Affairs (VHA) cohort,12 and models from the French prospective ANRS-CO12 Cir Vir cohort.13 In addition, 2 genetic prediction models for HCC were recently published,[14], [15], [16] drawing on common genetic polymorphisms, such as the rs738409 variant in the gene encoding Patatin-like phospholipase domain-containing protein 3 (PNPLA3).17
To enhance clinical decision-making, it is crucial that HCC prediction models are able to accurately predict HCC risk in a given patient. Inaccurate predictions have the potential to do harm. For example, underestimating HCC risk could lead to higher-risk patients being denied biannual ultrasound screening and, vice versa, overestimating HCC risk could lead to unnecessary screening in lower-risk patients.
At present, there are uncertainties regarding the performance of existing HCC prediction models. First, the acid-test of the accuracy of a prediction model is validation in a cohort that is independent from the one used to ‘train’ the model (known as external validation).18 Studies show that model performance is systematically better when measured on the same data set used to train the model vs. when measured on an ‘unseen’ dataset,19 Existing HCC prediction models have not been rigorously validated in external cohorts (with the exception of the aMAP10) and, thus, their performance could be overly optimistic. Second, studies have not adopted a competing risk perspective when evaluating model performance. This might be important because patients with cirrhosis are at high risk of dying from causes unrelated to HCC, such as liver failure20 and non-HCC cancer21; failing to take this into account could lead to biased estimates of prognosis.22 Third, the question of whether model performance is the same for all patients or whether it varies according to clinical characteristics, has not been explored. Fourth, genetic prediction models have practical advantages over non-genetic models (e.g. risk score is constant over time and, hence, only needs to be measured once). However, it is not clear how they compare performance-wise to their non-genetic counterparts. With these issues in mind, this study investigated the performance of selected HCC prediction models for patients with cured HCV cirrhosis in 2 separate UK cohorts.
Materials and methods
HCC prediction models
This study focuses on 6 HCC prediction models that are suitable or potentially suitable for patients with cirrhosis after HCV virological cure. The 6 models were: aMAP (2020)10; THRI (2018)11; VHA cirrhosis sustained virological response (SVR) score (2018)12; ANRS CO12 CirVir score (2017)13; Dongiovanni et al. genetic risk score (GRS; 2020)14,15; and Gellert-Kristensen et al. GRS (2020).16
For each prediction model, we extracted information from published articles relating to the following aspects of model derivation: sample size, average duration of follow-up, number of HCCs observed, proportion with HCV aetiology; specific prognostic factors selected, and the discrimination performance reported (Tables S1 and S2 and Appendix A).
Data sources for external validation
Model performance was assessed on patients with cirrhosis SVR from 2 UK cohorts, both followed from the date of SVR achievement.
STOP-HCV cohort
The STratified medicine to Optimise Treatment of Hepatitis C Virus (STOP-HCV) cirrhosis cohort is a prospective cohort study of 1,255 patients with liver cirrhosis and a history of chronic HCV infection. Participants were recruited from 31 liver clinics across the UK (except Northern Ireland) from January 2015 to July 2016 (i.e. coinciding with the introduction of direct-acting antivirals). Cirrhosis was defined on the basis of: (i) histological assessment (Ishak 5/6 or Metavir 4); or (ii) imaging results consistent with cirrhosis, including Fibroscan >15 kPa; or (iii) validated serum biomarker consistent with cirrhosis (including APRI >2 and Enhanced Liver Fibrosis [ELF®] test >10.48). Detailed clinical and laboratory information was collected on participants at the time of study enrolment and during subsequent annual study visits. Participants also provided a blood sample at enrolment, which was used to generate genotyping information using the Affymetrix UK Biobank array, which directly characterises individuals with respect to >800,000 genetic variants. Participants were also linked to health registries covering England and/or Wales, including the Hospital Episodes Statistics Admitted patients care dataset; cancer registrations collected by Public Health England; and death registrations. The study was approved by the West Midlands Research Ethics Committee (application reference: 14/WM/1128); Informed consent was obtained from all participants.
Scottish HCV clinical database
The Scottish HCV clinical database has been described extensively elsewhere.23,24 It is a retrospective cohort study of ∼25,000 patients in Scotland who have attended a specialist liver clinic appointment for the care and/or management of chronic HCV infection. The database records information collected during routine clinical care, including antiviral treatment episodes, diagnosis of cirrhosis, and the results of laboratory tests. It is also linked routinely to national health registries in Scotland, including the hospital, mortality, and cancer registers. Approval to link these registries and perform data analysis was granted by the Privacy Public Benefit Panel for Health and Social Care in NHS Scotland (application number: 1516-0457).
Liver cirrhosis was defined as compensated or decompensated cirrhosis diagnosed during routine clinical investigation. Diagnoses were typically made following liver biopsy, transient elastography, abdominal ultrasound, clinical examination, and routine liver function tests, according to clinical guidelines at that time. No information on genetic risk factors and/or polymorphisms was available in the Scottish cohort.
Inclusion criteria
For both cohorts, we included all patients with cirrhosis before initiating antiviral therapy and who subsequently achieved SVR. All SVRs were included irrespective of antiviral treatment regimen. If a patient had more than 1 treatment episode resulting in SVR, then the first episode was selected.
Exclusion criteria
Of those satisfying our aforementioned inclusion criteria, we then excluded patients as follows: for the STOP-HCV cohort, we first excluded participants recruited from Scottish and Welsh clinics for whom linkage data to health registries were unavailable or incomplete. This exclusion also ensured that there was no patient overlap between the STOP-HCV and Scottish cohorts. Second, we excluded participants with a diagnosis of HCC before completing antiviral therapy. Third, we excluded participants who had already achieved an SVR at the time of STOP-HCV study enrolment. This exclusion was applied to prevent immortal time bias25 between SVR achievement and STOP-HCV study enrolment. For the Scottish cohort, we excluded individuals with a history of HCC before achieving SVR. No other exclusions were made.
HCC risk predictions
Many of the prognostic factors included in the non-genetic HCC risk prediction models are dynamic insofar as they change over time. For prognostic factors based on laboratory tests (i.e. albumin, platelet count, etc.), we selected the most recent test on or before the start of antiviral treatment. Tests conducted more than 12 months before initiating treatment were excluded. We considered using laboratory tests conducted up to 12 months before SVR achievement to align with the date of follow-up commencement (see ‘Definition of risk sets’ section); however, we decided against this because antiviral treatment can cause acute and temporary changes in liver blood test values that might not necessarily reflect long-term risk profile. Nevertheless, age was based on age at the time of SVR achievement (i.e. time 0). Information on gamma glutamyl transferase was not available in the STOP-HCV cohort, precluding calculating values for the ANRS C012 CirVir model.13
Primary outcome event
The primary outcome event was diagnosis of HCC, identified through linkage to relevant administrative health databases. Specifically, for the STOP-HCV study, we used data from the England Admitted Patient Care Database, the National Cancer Registry, and Mortality Register to identify incident cases of HCC.
For the Scottish HCV clinical database, we used the equivalent National Inpatient Hospital Admission Database (SMR01), Cancer Registry (SMR6) and Mortality Register to identify HCC cases. For all registries/administrative databases, we used the standard ICD10 ‘C22.0’ or ICD9:155.0 code in the primary diagnostic and/or cause of death position to define HCC.
Statistical analysis
Definition of risk sets
All statistical analyses were underpinned by survival analysis methods. Follow-up time began at the date of SVR achievement. This was defined as 6 months after the treatment completion date for episodes initiated before the year 2014 (i.e. SVR24), and 3 months after the treatment completion date for episodes initiated from 2014 onwards (i.e. SVR12). This aligns with how SVR was defined by clinicians during the time period of this study. Follow-up ended at the date of incident HCC (if at all), mortality (if at all), or the date of study completion. For both cohorts, the study completion date was 1 January 2020, corresponding to the date the hospital admission registers were complete to. Unless indicated otherwise, non-HCC mortality was treated as a competing risk in all analyses.
Multiple imputation
Multiple imputation was used to replace missing data for the individual components of each HCC prediction model with plausible imputed estimates.26 We generated 20 imputations for each missing prediction using predictive mean matching. The following variables were used to predict these imputed values: (i) the Nelson–Aalen estimate of the baseline cumulative hazard; (ii) the outcome variable (i.e. HCC status); (iii) sex; (iv) decompensated cirrhosis; (v) age; (vi) alcohol use; and (vii) type of antiviral treatment [i.e. interferon (IFN) based or not]. Rubin’s rules were used to combine statistics of interest across imputation data sets.27 Similarly, cumulative incidence curves by risk tertile are based on the average estimate across the 20 imputation datasets created. Risk ‘tertiles’ refer to 3 groups: (i) those whose prediction is in the 33rd percentile or lower; (ii) those in the 33–67th percentile; and (iii) those in the 68–100th percentile.
Prediction model performance
Each prediction model was assessed in terms of 2 main aspects of prognostic model performance18: (i) discriminative ability (i.e. ability to differentiate between patients who develop HCC and those who do not); and (ii) calibration (agreement between the 3-year risk of HCC predicted by the model vs. the 3-year HCC risk observed).
Discrimination
The discriminative ability of each HCC prediction model was investigated in 3 ways.
First, we assessed the discrimination of each model visually, by plotting the cumulative incidence of HCC for individuals with low, moderate, and high scores. Categorisation into low, medium, and high groups was based on risk tertiles, as described earlier. Cumulative incidence was computed non-parametrically using the ‘stcompet’ command within Stata v16.28 Non-HCC mortality was treated as the competing risk event.
Second, we determined the discriminative ability of each prediction model quantitatively using the Concordance index (C-index), which provides an overall summary of the discriminative ability of a risk score. Specifically, the C-index measures the proportion of all possible ‘participant pairs’ that are ‘concordant’. A ‘participant pair’ refers to a random selection of 2 individuals from the data set, and this pair is said to be ‘concordant’ if the individual with the higher risk score develops the outcome event of interest sooner than the individual with the lower risk score.29 In our base-case analysis, we used a version of the C-index adapted for a competing-risk scenario, as previously described by Wolbers et al.30 The key difference between the standard C-index and competing-risk adjusted C-index is that, in the latter, individuals with a competing risk event are assumed to have an infinite survival time. In addition, we also calculated the standard Harrell C-index, which does not account for competing risks. For all versions of the C-index, higher values indicate better discrimination; a value of 0.5 indicates zero discrimination (i.e. no better than chance), whereas a value of 1.0 indicates perfect discrimination.
Third, we assessed whether the C-index of each prediction model varied according to selected patient characteristics. These characteristics were as follows: age <60 years; sex; history of heavy alcohol use (defined as consumption of >50 units/week for a sustained period of >6 months before SVR); genotype 3; and SVR through IFN-free therapy.
Calibration
Calibration measures how closely the predicted risk of HCC matches the observed risk of HCC.18,31 For this analysis, we calculated the 3-year predicted and 3-year observed risk of HCC for individuals with low, moderate, and high predictions (again defined according to risk tertiles).
The predicted 3-year probability of HCC was calculated using standard Cox regression, as prescribed by the authors of each risk score, namely:
| 1–S0(t)exp(linear predictor) |
where t = 3 years, and S0(t) refers to the estimated 3-year HCC-free survival for individuals with zero for all independent variables in the model. We contacted the authors for this information if these details were not clear in the original paper.
Our calculation of the 3-year observed HCC probability was based on the cumulative incidence function, with non-HCC mortality treated as a competing risk.
Finally, we did not perform a calibration analysis for the genetic models, because they were not intended to estimate the probability of HCC at a particular point in time.
Results
Derivation of external validation cohorts
In total, 2,245 patients met our inclusion criteria from the Scottish cohort. We then excluded 106 patients with HCC before treatment completion. Thus, the final sample size was 2,139 (Fig. S1).
Overall, 1,019 participants from the STOP-HCV study met our inclusion criteria. We then excluded 79 patients from Scotland and Wales. In addition, 77 patients with HCC before SVR achievement were also excluded. Finally, a further 257 patients who achieved SVR before enrolling in STOP-HCV were removed to avoid immortal time bias. Thus, the final sample size was 606 (Fig. S1).
Patient characteristics
Patients in both cohorts were mainly middle-aged (i.e. between 40 and 65 years old), male (>70%), and of white ethnicity (>80%) (Table 1). However, there were notable differences between these 2 cohorts. First, patients in the STOP-HCV cohort were older than in the Scottish cohort (mean age: 56.5 vs. 50.2 years, respectively). Second, the proportion of patients who had achieved SVR through IFN-free therapies was higher in the STOP-HCV that in the Scottish cohort (92% achieved SVR via IFN-free therapies in STOP-HCV vs. 61% in Scottish cohort). Third, the proportion of patients with past HCV genotype 3 infection was lower in the STOP-HCV cohort (38% vs. 50%, respectively). Finally, average values for the VHA, THRI, and aMAP scores were all higher in the STOP-HCV cohort vs. the Scottish cohort, indicating that STOP-HCV had higher predicted HCC risk.
Table 1.
Description of Scottish and STOP-HCV cohorts.
| Characteristic | Scottish cohort (n = 2,139) |
STOP-HCV cohort (n = 606) |
||
|---|---|---|---|---|
| Mean value/proportion | Number with missing data (%) | Mean value/proportion/allele frequency | Number with missing data (%) | |
| Demographic, clinical, and behavioural factors | ||||
| Age, years (SD) | 50.2 (9.0) | 0 (0.0) | 56.5 (9.6) | 0 (0.0) |
| % Age >60 years | 14.0 | 0 (0.0) | 38.4 | 0 (0.0) |
| % Male sex | 74.0 | 0 (0.0) | 70.6 | 0 (0.0) |
| % White ethnicity | 94.3 | 0 (0.0) | 82.0 | 0 (0.0) |
| % IFN-free therapy | 61.1 | 0 (0.0) | 91.7 | 0 (0.0) |
| % Decompensated cirrhosis | 10.5 | 0 (0.0) | 11.2 | 0 (0.0) |
| % Past genotype 3 infection | 50.1 | 21 (1.0) | 38.3 | 34 (5.6) |
| % IDU history | 75.7 | 379 (17.7) | 44.8 | 30 (5.0) |
| Laboratory markers | ||||
| ALBI (SD) | -2.43 (0.53) | 297 (13.9) | -2.62 (0.54) | 55 (9.1) |
| Platelet count (SD) | 148.4 (68.0) | 271 (12.7) | 136.3 (66.7) | 57 (9.4) |
| ALT (SD) | 88.0 (71.5) | 253 (11.8) | 90.2 (65.6) | 61 (10.1) |
| AST (SD) | 84.3 (56.8) | 443 (20.7) | 89.0 (59.3) | 106 (17.5) |
| GGT (SD) | 154.6 (172.4) | 1,034 (48.3) | Not available | n.a. |
| Albumin (SD) | 37.2 (5.2) | 296 (13.8) | 39.4 (5.3) | 54 (8.9) |
| Genetic markers | ||||
| rs738309:G AF | Not available | n.a. | 26.1 | 60 (9.9) |
| rs58542926:T AF | 8.2 | 60 (9.9) | ||
| rs72613567:TA AF | 20.3 | 67 (11.1) | ||
| rs641738:T AF | 41.1 | 60 (9.9) | ||
| rs1260326:T AF | 37.6 | 60 (9.9) | ||
| HCC prediction model scores | ||||
| aMAP (SD) | 57.1 (7.4) | 336 (15.7) | 59.5 (7.2) | 64 (10.6) |
| VHA model (SD) | 0.64 (0.47) | 510 (23.8) | 0.88 (0.55) | 123 (20.3) |
| THRI model (SD) | 145.9 (58.6) | 271 (12.7) | 168.4 (58.8) | 62 (10.2) |
| ANRS CO12 CirVir (SD) | 4.4 (2.0) | 1,288 (60.2) | Not available | – |
| Gellert-Kristensen GRS (SD) | n.a. | n.a. | 2.28 (0.91) | 67 (11.1) |
| Dongiovanni GRS (SD) | n.a. | n.a. | 0.28 (0.21) | 60 (9.9) |
HCC prediction model scores refer to the raw values and are all on different scales. Laboratory markers are based on values at the time of treatment initiation, whereas all other dynamic variables (e.g. age) are based on the value at SVR achievement. See main text for further explanation.
AF, allele frequency; ALBI, albumin/bilirubin; ALT, alanine aminotransferase; aMAP, age-male sex-ALBI-platelet count score; AST, aspartate aminotransferase; GGT, gamma glutamyl transferase; GRS, genetic risk score; HCC, hepatocellular carcinoma; IDU, injecting-drug user; IFN, interferon; STOP-HCV, STratified medicine to Optimise Treatment of Hepatitis C Virus; THRI, Toronto HCC Risk Index; VHA, Veteran Health Affairs.
The proportion of patients with missing predictions was generally <20%. However, missing data were more substantial in the Scottish cohort for the VHA model (24% missing) and the ANRS C012 model (60% missing) compared with the other models.
Cumulative incidence of HCC and non-HCC mortality
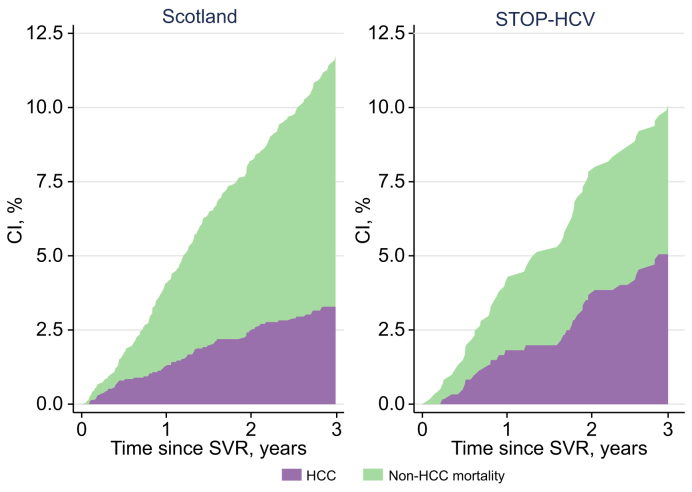
In the Scottish cohort, participants were followed for a mean 3.9 years after SVR, during which time 118 incident HCC events and 214 non-HCC-related deaths occurred (Table 2). The cumulative incidence of HCC and non-HCC mortality at 3 years was 3.3% (95% CI 2.6–4.2) and 8.5% (95% CI 7.2–9.8), respectively (Table 2 and Fig. 1).
Table 2.
Description of follow-up data and outcome events observed in the Scottish and STOP-HCV cohorts.
| Cohort | No. of individuals | Person years (PYs) |
Outcome |
|||||
|---|---|---|---|---|---|---|---|---|
| Total | Mean per patient | Median per patient | Event | No. of events | Crude rate, per 1,000 PYs (95% CI) | 3-year cumulative incidence (%) | ||
| Scottish cohort | 2,139 | 8,380 | 3.9 | 3.5 | HCC | 118 | 14.1 (11.8–16.9) | 3.3% (2.6–4.2) |
| Non-HCC mortality | 214 | 25.5 (22.3–29.2) | 8.5% (7.2–9.8) | |||||
| Drug-related mortality | 52 | 6.2 (4.7–8.1) | 2.2% (1.6–2.9) | |||||
| External causes mortality | 12 | 1.4 (0.8–2.5) | 0.6% (0.3–1.0) | |||||
| Non-HCC liver mortality | 45 | 5.4 (4.1–7.2) | 2.1% (1.5–2.8) | |||||
| All-cause mortality | 278 | 32.2 (28.6–36.2) | 9.8% (8.5–11.2) | |||||
| STOP-HCV cohort | 606 | 2,041 | 3.4 | 3.7 | HCC | 40 | 19.60 (14.4–26.7) | 5.1% (3.5–7.0) |
| Non-HCC mortality | 36 | 17.6 (12.7–24.5) | 5.0% (3.5–7.0) | |||||
| Drug-related mortality | 3 | 1.5 (0.5–4.6) | 0.5% (0.1–1.4) | |||||
| External causes mortality | 0 | 0 | 0 | |||||
| Non-HCC liver mortality | 18 | 8.8 (5.6–14.0) | 2.2% (1.2–3.6) | |||||
| All-cause mortality | 50 | 23.9 (18.1–31.6) | 7.3% (5.4–9.6) | |||||
Drug-related, external causes, and non-HCC liver mortality represent specific types of non-HCC mortality.
HCC, hepatocellular carcinoma; STOP-HCV, STratified medicine to Optimise Treatment of Hepatitis C Virus.
Fig. 1.
Stacked cumulative incidence curves for HCC and non-HCC mortality.
Cumulative incidence curves are generated non-parametrically (i.e. without any modelling assumptions). For the purple line, non-HCC mortality is treated as a competing risk event, whereas for the green line, HCC outcome is treated as a competing risk event. CI, cumulative incidence; HCC, hepatocellular carcinoma.
Patients in the STOP-HCV cohort were followed for a mean of 3.4 years after SVR, during which time 40 incident HCCs and 36 non-HCC deaths occurred (Table 2). The cumulative incidences of HCC and non-HCC mortality at 3 years were 5.1% (95% CI 3.5–7.0) and 5.0% (95% CI 3.5–7.0), respectively (Fig. 1).
Drug-related mortality and deaths from external causes were more common in the Scottish cohort vs. the STOP-HCV cohort. One-third of non-HCC mortality was from drug-related or external causes in the Scottish cohort, compared with only 10% in the STOP-HCV study (Table 2).
Performance of HCC prediction models
Discrimination
In cumulative incidence plots, higher predicted HCC risks were associated with a higher HCC cumulative incidence (Figs S2 and S3). However, the degree of discrimination varied considerably by both prediction model and cohort.
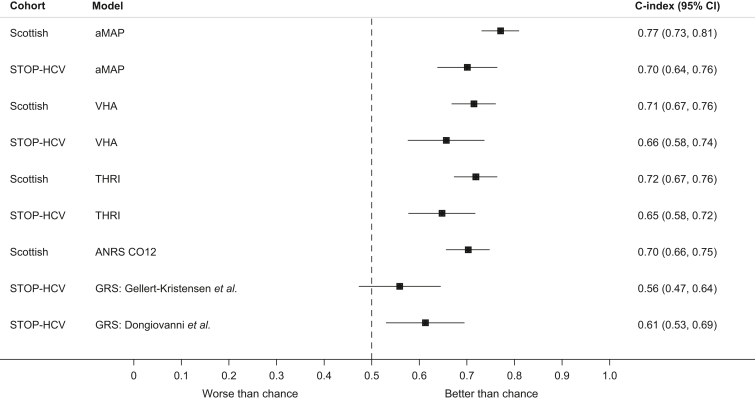
In the Scottish cohort, the aMAP score exhibited the best discrimination (C-index: 0.771; 95% CI 0.731–0.810), followed by the VHA model (0.715; 95% CI 0.668–0.761), THRI (0.719; 95% CI 0.673–0.764), and ANRS CO12 (0.703; 95% CI 0.656–.749) (Fig. 2).
Fig. 2.
Discriminative ability of HCC prediction models in Scottish and STratified medicine to Optimise Treatment of Hepatitis C Virus (STOP-HCV) cohorts in terms of the Concordance index (C-index).
C-index refers specifically to the Wolbers Concordance index, which takes account of competing risk events. Here, non-HCC mortality is treated as a competing risk. The dashed line represents the point of zero discrimination. aMAP, age-male sex-ALBI-platelet count score; GRS, genetic risk score; HCC, hepatocellular carcinoma; THRI, Toronto HCC Risk Index; VHA, Veteran Health Affairs.
HCC prediction models exhibited poorer discriminative performance (i.e. lower C-index values) in the STOP-HCV cohort, but the general ranking was similar. For example, aMAP was also the top-performing score in the STOP-HCV cohort (C-index: 0.701; 95% CI 0.638–0.764), followed by VHA (0.657; 95% CI 0.576–0.737), and then by THRI (0.648; 95% CI 0.577–0.718). The Dongiovanni GRS had a C-index value of 0.613 (95% CI 0.530–0.695) and the Gellert-Kristensen GRS C-index value was 0.559 (95% CI 0.473–0.645) (Fig. 2). All C-index values were marginally higher when using the standard Harrell’s C-index as opposed to the Wolbers-modified C-index (Table S3).
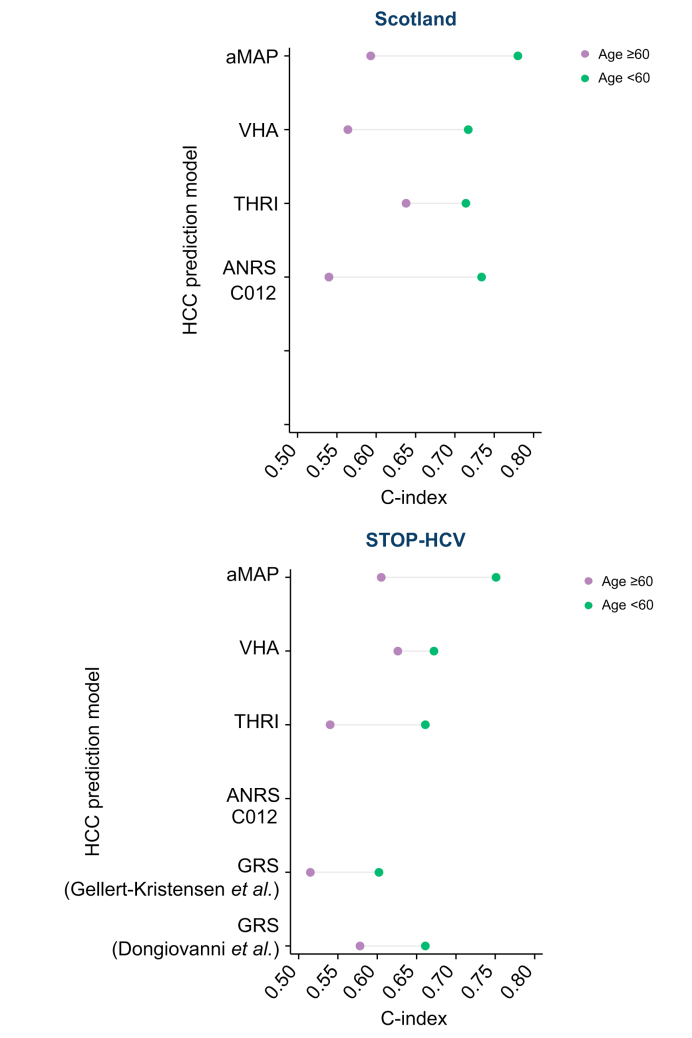
Variability in discrimination
Our analysis of variability in model discrimination highlighted 2 patient factors of interest (Figs S4 and S5). First, discrimination was better for younger patients vs. older patients. This was apparent across both cohorts for all non-genetic models. For example, in the Scottish cohort, the aMAP had a C-index of 0.59 (95% CI 0.49–0.70) for those aged >60 years at SVR achievement vs. 0.80 (95% CI 0.75–0.84) for those aged <60 years (Fig. 3). Second, GRS discrimination was better for patients with past genotype 3 infection. For example, the C-index of the Dongiovanni GRS was 0.78 (95% CI 0.70–0.87) in patients with genotype 3 vs. 0.50 (95% CI 0.39–0.62) in patients with non-genotype 3.
Fig. 3.
Comparison of the discriminative ability for HCC incidence, according to age, based on the Wolbers Concordance index, taking account of non-HCC mortality as a competing risk in the Scottish and STOP-HCV cohorts.
aMAP, age-male sex-ALBI-platelet count score; GRS, genetic risk score; HCC, hepatocellular carcinoma; THRI, Toronto HCC Risk Index; VHA, Veteran Health Affairs.
Otherwise, no major heterogeneity in model discrimination was observed according to IFN-free therapies, alcohol history, sex, or decompensated disease. (Figs S4 and S5).
Calibration
In the Scottish cohort, the observed 3-year probability of HCC was 3.3% (95% CI 2.6–4.2), compared with predicted probabilities of 2.0% (THRI), 3.1% (VHA), 3.6% (aMAP), and 3.9% (ANRS CO12 model). In STOP-HCV, the 3-year observed probability of HCC was 5.1% (95% CI 3.5–7.0) compared with predicted probabilities of 2.5% (THRI model), 3.8% (VHA model), and 5.0% (aMAP model) (Fig. S6).
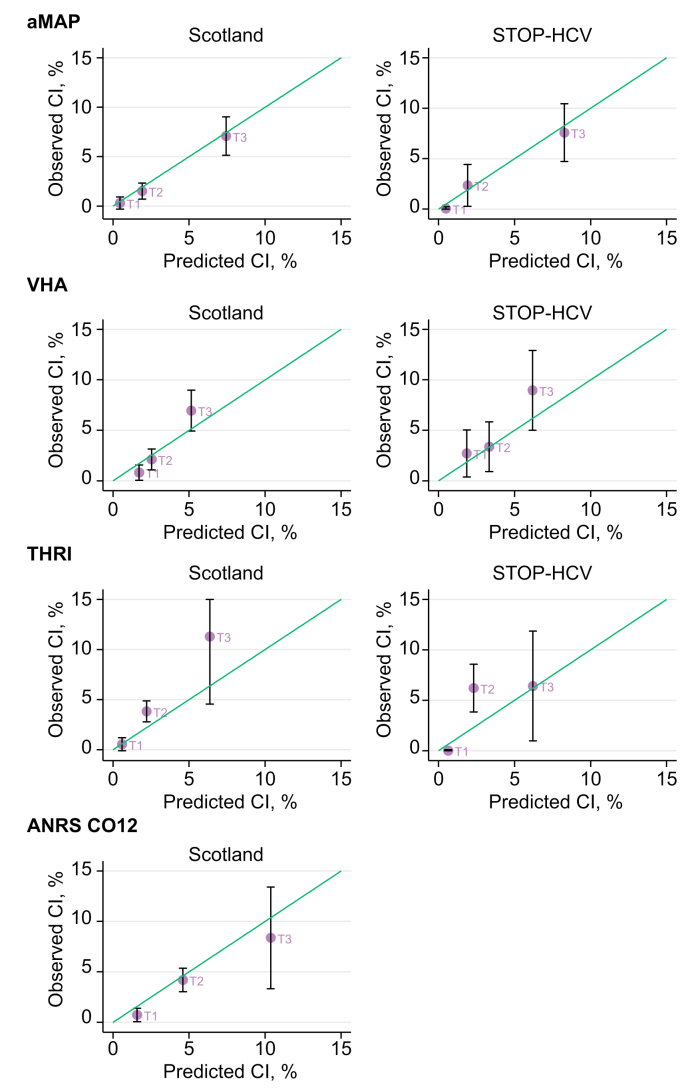
When we examined calibration according to risk tertiles, we saw some instances of underprediction in higher-risk patients. For example, in the Scottish cohort, the observed 3-year risk for individuals whose THRI score was in tertile 3 (11.3% 95% CI 4.6–18.0) was almost twice the predicted risk (6.4%). This underprediction also affected the VHA model to some extent, but did not affect either the aMAP or ANRS CO12 models (Fig. 4).
Fig. 4.
Agreement between observed and predicted 3-year HCC probability, by risk tertile.
T1, T2, and T3 denote risk tertiles 1, 2, and 3, respectively. Risk tertiles refer to 3 groups: (i) T1, those whose prediction is in the 33rd percentile or lower; (ii) T2, those in the 33rd to 67th percentile; and (iii) T3: those in the 68th to 100th percentile. The green line indicates perfect agreement between observed and predicted risk. Values above the green line indicate that the observed risk is higher than the predicted risk (and vice versa). aMAP, age-male sex-ALBI-platelet count score; CI, cumulative incidence; HCC, hepatocellular carcinoma; STOP-HCV, STratified medicine to Optimise Treatment of Hepatitis C Virus; THRI, Toronto HCC Risk Index; VHA, Veteran Health Affairs.
Discussion
HCC risk prediction models have the potential to support clinical decision-making, but could equally cause harm if their predictions are not robust. In this study, we used external validation to quantify the performance of existing HCC prediction models for individuals with cirrhosis and cured HCV, with 3 key findings. First, our data confirm that HCC prediction models are able to discriminate between patients who go on to develop HCC and those who do not. In other words, across all models, an increase in predicted HCC risk was mirrored by an increase in observed HCC risk (and vice versa). Nevertheless, not all models provide the same level of discrimination in a UK setting. Overall, the aMAP model exhibited the best discriminative ability, with a C-index of 0.78 in the Scottish and 0.71 in the STOP-HCV cohorts. The aMAP model is derived entirely from routinely available prognostic factors (i.e. age, albumin, bilirubin, platelet count, and sex) and, thus, this provides encouragement regarding opportunities for ‘real-world’ risk stratification in this growing patient group. Another corollary is that aMAP should be used as a benchmark for rival prognostic models to surpass. This will help the research community evaluate whether a proposed new model (of which many are likely to emerge in the years ahead) provides added value over existing alternatives. A second novel aspect of this study is that it highlights the existence of heterogeneity in model performance (i.e. variability in model performance according to patient characteristics). For example, we found that most prediction models were more discriminating in younger patients vs. older patients (Fig. 3), although it is unclear why this is the case. In a similar vein, we showed that the Dongiovanni et al. GRS exhibited better discrimination for patients with past genotype 3 infection (C-index: 0.78) than for those with non-genotype 3 infection (C-index: 0.50). This could be because the Dongiovanni GRS was originally developed as a risk score for hepatic steatosis, which is well known to be a more prominent histological feature of HCV genotype 3 infection vs. genotype 2/3 infection.32,33 A third important observation from this study is to caution that some models might underpredict 3-year HCC risk. This was most prominent for the THRI model among higher-risk patients. Thus, recalibration might be necessary before adopting this model in a UK setting (although we acknowledge that underpredicting high-risk patients would probably not alter HCC screening decisions).
An important question that this study does not answer is how to translate a HCC risk prediction into an HCC screening decision for a given patient. There is agreement in the field that HCC risk prediction models will have most clinical utility for identifying patients whose risk of HCC is too low for screening to be of net benefit. However, there is considerable ambiguity regarding which patients are ‘low risk’ and how this should be defined. In our view, the definition of low risk should reflect a compromise between multiple factors, such as: (i) cost-effectiveness data; (ii) general population HCC incidence; (iii) patient preferences; (iv) clinical view and other clinical factors (e.g. likelihood of receiving curative treatment in the event of a HCC diagnosis); and (v) resources available for HCC screening. In the real world, ‘low risk’ is likely to represent a range of values rather than a hard threshold, and is unlikely to be the same for all patients. It will also inevitably change as new surveillance technologies emerge with different performance characteristics to abdominal ultrasound. Thus, to support HCC screening decisions, versatile models are needed with good calibration across the risk spectrum. This is why we focused on calibration in this study. We deliberately avoided defining ‘low risk’ based on what is optimal for a given model (i.e. which previous studies have done by identifying the risk threshold at which the sensitivity/specificity are optimised). This approach is statistically dubious,34 but more to the point, it is equivalent to letting a statistical model dictate a clinical decision, as opposed to using a statistical model to help implement a clinical decision. Thus, auxiliary research to define ‘low risk’ might be needed before models, such as aMAP, can be confidently deployed. Microsimulation Markov models35 could be useful for estimating the benefits of screening (i.e. in terms of life-years or quality-adjusted-life-years gained) according to 3-year HCC probability.
This study has several strengths. First, our focus on externally validating competing models fills an important gap in the literature (i.e. most previous studies have opted to develop new risk models, rather than evaluate the performance of existing ones). Second, as previously discussed, we assessed model performance in terms of not only discrimination, but also calibration. A third strength is that our estimates of model performance account for non-HCC mortality as a competing risk. This perspective is important because patients with cirrhosis are at high risk of mortality from liver failure, and this can bias estimates of model performance.22 However, although we found that C-indexes were lower when accounting for competing risks, the differences were modest. A fourth strength is the adoption of a dual-cohort perspective, enabling us to perform the same analysis in 2 different cohorts and analyse variability. This supported our investigation of heterogeneity in model performance. Another unique asset of this study is that we collected data on genetic and non-genetic models and, thus, were able to compare the discriminative ability of these 2 model types. Our study also has limitations that warrant discussion. One of the main limitations is that predictions were missing for some patients. Although the proportion of missing data was generally low (<20%), missing data were more substantial for the ANRS-CO12 (60% missing from the Scottish cohort) and VHA models (24% missing from the Scottish cohort). We used multiple imputation to maximise statistical power and correct potential bias from a complete-case analysis. Nevertheless, the performance of the ANRS-CO12 model in particular should be viewed with caution in light of the missing data. Second, we cannot exclude the possibility that some of the patients in our data set might have had HCC or been developing HCC before SVR was achieved. Third, we were unable to evaluate all models developed so far for patients with HCV cirrhosis, including those proposed by Pons et al.,36 Audurea et al.,37 and Alonso Lopez et al.38 These scores were omitted from our analysis because data for factors such as liver stiffness and prothrombin time were unavailable in the Scottish and STOP-HCV studies. This is also an inherent weakness of the scores themselves insofar as a model can only be useful if it can be calculated using ‘real-world’ data. Fourth, although patients were followed up from the point of SVR achievement, we did not have information on the specific date that the SVR test was performed. Thus, a conservative estimate of 6 months after treatment completion was used to ensure our analysis was not affected by immortal time bias25 (i.e. equivalent to SVR24).
In summary, this is the first study comparing the performance of competing HCC risk prediction models. Our findings highlight the opportunities for practical HCC risk stratification in a UK setting for patients with cirrhosis and cured HCV. However, if models are to support HCC screening decisions, then a consensus will ultimately be needed regarding the individualised probability of HCC at which screening should be avoided.
Financial support
This study received financial support from the Medical Research Foundation (Grant ID: C0825), to establish HCV Research UK; Public Health Scotland; the Medical Research Council (Grant ID: MR/K01532X/1), which funded the STOP-HCV study; the Royal Society and Wellcome Trust (220171/Z/20/Z to M.A.A.); and Cancer Research UK (C30358/A29725). H.I. is supported by a viral hepatitis fellowship from the Medical Research Foundation (award no: C0825).
Authors’ contributions
Study concept: all authors. Study design: HI, JP, SH, ING, WI, EB, SM. Acquisition of data: HI, WI, SH, ING, EB. Resources: all authors. Statistical analysis: HI, JP, SM. Drafting manuscript: HI, JP. Critical revision of manuscript: all authors.
Data availability statement
The Scottish data used in this study are not publicly available, but can be acquired through successful application to the Public Benefit and Privacy Panel for Health and Social Care (www.informationgovernance.scot.nhs.uk/pbpphsc/home/for-applicants/). The STOP-HCV consortium welcomes collaboration with interested parties. Anonymised samples and clinical data held on the study database are accessible through application to the HCVRUK Tissue Data Access Committee. Contact Professor Indra Neil Guha (neil.guha@nottingham.ac.uk) or Professor William Irving (will.irving@nottingham.ac.uk) for more information. Please note that we cannot share linked NHS digital data with other research groups, or any data variables derived from linked NHS digital data.
Conflicts of interest
There are no conflicts of interest to disclose.
Please refer to the accompanying ICMJE disclosure forms for further details.
Acknowledgements
We would like to thank the participants of the STOP-HCV cirrhosis study. HCV Research UK participating sites and Principal Investigators are as follows: K. Agarwal (Kings College Hospital, London), M. Aldersley (St James’s University Hospital, Leeds), R. Aspinall (Queen Alexandra Hospital, Portsmouth), S. Barclay (Glasgow Royal Infirmary), E. Barnes (John Radcliffe Hospital, Oxford), J. Benselin (University of Nottingham), A. Brown (St Mary’s Hospital, London), C. Ch’ng (Singleton Hospital, Swansea), L. Corless (Hull Royal Infirmary), M. Cramp (Derriford Hospital, Plymouth), J. Dillon (Ninewells Hospital, Dundee), Shirley English (Aberdeen Royal Infirmary); D. Forton (St George’s Hospital, London), G. Foster (The London Hospital), A. Fraser (Aberdeen Royal Infirmary), W. Gelson (Addenbrookes Hospital, Cambridge), D. Gorard (Wycombe Hospital), F. Gordon (Bristol Royal Infirmary), N. Kennedy (Monklands Hospital), J. Knowles (James Cook University Hospital, Middlesbrough), C. Leen (Western General Hospital, Edinburgh S. McPherson (Freeman Hospital, Newcastle), S. Moreea (Bradford Teaching Hospitals NHS Foundation Trust), D. Mutimer (Queen Elizabeth Hospital, Birmingham), M. Prince (Manchester Royal Infirmary, P. Richardson (Royal Liverpool University Hospital), W. Rosenberg (Royal Free Hospital & University College Hospital), S. Ryder (Queen’s Medical Centre, Nottingham), K. Rye (Royal Shrewsbury Hospital), B. Stone (Royal Hallamshire Hospital, Sheffield), M. Thursz (St Mary’s Hospital, London), A. Ustianowski (North Manchester General Hospital), S. Verma (Royal Sussex County Hospital, Brighton) and M. Wiselka (Leicester Royal Infirmary). The STOP-HCV Consortium comprises: E. Barnes (University of Oxford), J.K. Ball (University of Nottingham), D. Brainard (Gilead Sciences), G. Burgess (Conatus Pharmaceuticals), G. Cooke (Imperial College, London), J. Dillon (University of Dundee), G.R. Foster (Queen Mary University of London), C. Gore (Hepatitis C Trust), N. Guha (University of Nottingham), R. Halford (Hepatitis C Trust), K. Whitby (Gilead Sciences), C. Holmes (University of Oxford), A. Howe (British Columbia Centre for Excellence), E. Hudson (University of Oxford), S. Hutchinson (Glasgow Caledonian University), W.L. Irving (University of Nottingham), S. Khakoo (University of Southampton), P. Klenerman (University of Oxford), N. Martin (UC San Diego), B. Massetto (Gilead Sciences), T. Mbisa (Public Health England), J. McHutchinson (Gilead Sciences), J. McKeating (University of Oxford), J. McLauchlan (Centre for Virus Research, Glasgow), A. Miners (London School of Hygiene and Tropical Medicine), A. Murray (OncImmune Limited), P. Shaw (Merck & Co), P. Simmonds (University of Oxford), C. Spencer (Wellcome Trust), E. Thomson (Centre for Virus Research, Glasgow), P. Vickerman (University of Bristol) and N. Zitzmann (University of Oxford). For the purpose of open access, the author has applied a CC BY public copyright licence to any Author Accepted Manuscript version arising from this submission. The views expressed in this article are those of the authors and not necessarily those of the NHS, NIHR, or Department of Health.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhepr.2021.100384.
Supplementary data
The following are the supplementary data to this article:
References
- 1.El-Serag H.B., Kanwal F., Richardson P., Kramer J. Risk of hepatocellular carcinoma after sustained virological response in veterans with hepatitis C virus infection. Hepatology. 2016;64:130–137. doi: 10.1002/hep.28535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Innes H., Barclay S.T., Hayes P.C., Fraser A., Dillon J.F., Stanley A., et al. The risk of hepatocellular carcinoma in cirrhotic patients with hepatitis C and sustained viral response: role of the treatment regimen. J Hepatol. 2018;68:646–654. doi: 10.1016/j.jhep.2017.10.033. [DOI] [PubMed] [Google Scholar]
- 3.van der Meer A.J., Feld J.J., Hofer H., Almasio P.L., Calvaruso V., Fernandez-Rodriguez C.M., et al. Risk of cirrhosis-related complications in patients with advanced fibrosis following hepatitis C virus eradication. J Hepatol. 2017;66:485–493. doi: 10.1016/j.jhep.2016.10.017. [DOI] [PubMed] [Google Scholar]
- 4.Ioannou G.N., Beste L.A., Green P.K., Singal A.G., Tapper E.B., Waljee A.K., et al. Increased risk for hepatocellular carcinoma persists up to 10 years after HCV eradication in patients with baseline cirrhosis or high FIB-4 scores. Gastroenterology. 2019;157:1264–1278. doi: 10.1053/j.gastro.2019.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allemani C., Matsuda T., Di Carlo V., Harewood R., Matz M., Niksic M., et al. Global surveillance of trends in cancer survival 2000-2014 (CONCORD-3): analysis of individual records for 37 513025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet. 2018;391:1023–1075. doi: 10.1016/S0140-6736(17)33326-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2012;56:908–943. doi: 10.1016/j.jhep.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 7.Heimbach J.K., Kulik L.M., Finn R.S., Sirlin C.B., Abecassis M.M., Roberts L.R., et al. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology. 2018;67:358–380. doi: 10.1002/hep.29086. [DOI] [PubMed] [Google Scholar]
- 8.Kanwal F., Singal A.G. Surveillance for hepatocellular carcinoma: current best practice and future direction. Gastroenterology. 2019;157:54–64. doi: 10.1053/j.gastro.2019.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujiwara N., Friedman S.L., Goossens N., Hoshida Y. Risk factors and prevention of hepatocellular carcinoma in the era of precision medicine. J Hepatol. 2018;68:526–549. doi: 10.1016/j.jhep.2017.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan R., Papatheodoridis G., Sun J., Innes H., Toyoda H., Xie Q., et al. aMAP risk score predicts hepatocellular carcinoma development in patients with chronic hepatitis. J Hepatol. 2020;73:1368–1378. doi: 10.1016/j.jhep.2020.07.025. [DOI] [PubMed] [Google Scholar]
- 11.Sharma S.A., Kowgier M., Hansen B.E., Brouwer W.P., Maan R., Wong D., et al. Toronto HCC risk index: a validated scoring system to predict 10-year risk of HCC in patients with cirrhosis. J Hepatol. 2018;68:92–99. doi: 10.1016/j.jhep.2017.07.033. [DOI] [PubMed] [Google Scholar]
- 12.Ioannou G.N., Green P.K., Beste L.A., Mun E.J., Kerr K.F., Berry K. Development of models estimating the risk of hepatocellular carcinoma after antiviral treatment for hepatitis C. J Hepatol. 2018;69:1088–1098. doi: 10.1016/j.jhep.2018.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ganne-Carrie N., Layese R., Bourcier V., Cagnot C., Marcellin P., Guyader D., et al. Nomogram for individualised prediction of hepatocellular carcinoma occurrence in hepatitis C virus cirrhosis (ANRS CO12 CirVir) Hepatology. 2016;64:1136–1147. doi: 10.1002/hep.28702. [DOI] [PubMed] [Google Scholar]
- 14.Dongiovanni P., Stender S., Pietrelli A., Mancina R.M., Cespiati A., Petta S., et al. Causal relationship of hepatic fat with liver damage and inuslin resistance in non-alcoholic fatty liver. J Intern Med. 2018;283:356–370. doi: 10.1111/joim.12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Degasperi E., Galmozzi E., Pelusi S., D’ambrosio R., Soffredini R., Borghi M., et al. Hepatic fat – genetic risk score predicts hepatocellular carcinoma in patients with cirrhotic HCV treated with DAAs. Hepatology. 2020;72:1912–1923. doi: 10.1002/hep.31500. [DOI] [PubMed] [Google Scholar]
- 16.Gellert-Kristensen H., Richardson T.G., Davey-Smith G., Nordestgaard B.G., Tybjaerg-Hansen A., Stender S. Combined effect of PNPLA3, TM6SF2 and HSD17B13 variants on risk of cirrhosis and hepatocellular carcinoma in the general population. Hepatology. 2020;72:845–856. doi: 10.1002/hep.31238. [DOI] [PubMed] [Google Scholar]
- 17.Trepo E., Romeo S., Zucman-Rossi J., Nahon P. PNPLA3 gene in liver diseases. J Hepatol. 2016:399–412. doi: 10.1016/j.jhep.2016.03.011. [DOI] [PubMed] [Google Scholar]
- 18.Moons K.G.M., Altman D.G., Reitsma J.B., Ioannidis J.P.A., Macaskill P., Steyerberg E.W., et al. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): explanation and elaboration. Ann Intern Med. 2015;162:W1–W73. doi: 10.7326/M14-0698. [DOI] [PubMed] [Google Scholar]
- 19.Collin G.S., de Groot J.A., Dutton S., Omar O., Shanyinde M., Tajar A., et al. External validation of multivariable prediction models: a systematic review of methodological conduct and reporting. BMC Med Res Methodol. 2014;14:40. doi: 10.1186/1471-2288-14-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D’Amico G., Garcia-Tsao G., Pagliaro L. Natural history and prognostic indicators of survival in cirrhosis: a systematic review of 118 studies. J Hepatol. 2006;44:217–231. doi: 10.1016/j.jhep.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 21.Hong C.Y., Sinn D.H., Kang D., Paik S.W., Guallar E., Cho J., et al. Incidence of extrahepatic cancers among individuals with chronic hepatitis B or C virus infection: a nationwide cohort study. J Viral Hepat. 2020;27:896–903. doi: 10.1111/jvh.13304. [DOI] [PubMed] [Google Scholar]
- 22.Jepsen P., Vilstrup V., Andersen P.K. The clinical course of cirrhosis: the importance of multistate models and competing risks analysis. Hepatology. 2015;62 doi: 10.1002/hep.27598. 2922–302. [DOI] [PubMed] [Google Scholar]
- 23.McDonald S.A., Hutchinson S.J., Innes H.A., Allen S., Bramley P., Bhattacharyya, et al. Attendance at specialist hepatitis clinics and initiation of antiviral treatment among persons chronically infected with hepatitis C: examining the early impact of Scotland’s hepatitis C action plan. J Viral Hepat. 2014;21:366–376. doi: 10.1111/jvh.12153. [DOI] [PubMed] [Google Scholar]
- 24.McDonald S.A., Innes H.A., Hayes P.C., Dillon J.F., Mills P.R., Goldberg D.J., et al. What is the impact of a country-wide scale-up in antiviral therapy on the characteristics and sustained viral response rates of patients treated for hepatitis C? J Hepatol. 2015;62:262–268. doi: 10.1016/j.jhep.2014.08.046. [DOI] [PubMed] [Google Scholar]
- 25.Suissa S. Immortal time bias in pharmaco-epidemiology. Am J Epidemiol. 2008;167:492–499. doi: 10.1093/aje/kwm324. [DOI] [PubMed] [Google Scholar]
- 26.Sterne J.A.C., White I.R., Carlin J.B., Spratt M., Royston P., Kenward M.G., et al. Multiple imputation for missing data in epidemiological and clinical research: potential and pitfalls. BMJ. 2009;338:B2393. doi: 10.1136/bmj.b2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rubin D. Wiley; New York: 1987. Multiple Imputation for Nonresponse in Surveys. [Google Scholar]
- 28.Coviello E. Boston College Department of Economics; Boston: 2012. STCOMPET: Stata Module to Generate Cumulative Incidence in Presence of Competing Events: Statistical Software Components S431301. [Google Scholar]
- 29.Pencina M.J., D’Agostino R.B., Song L. Quantifying discrimination of Framingham risk functions with different survival C statistics. Stat Med. 2012;31:1543–1553. doi: 10.1002/sim.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wolbers M., Koller M.T., Witteman J.C.M., Steyerberg E.W. Prognostic models with competing risks. methods and application to coronary risk prediction. Epidemiology. 2009;20:555–561. doi: 10.1097/EDE.0b013e3181a39056. [DOI] [PubMed] [Google Scholar]
- 31.Royston P., Altman D.G. External validation of a Cox prognostic model: principles and methods. BMC Med Res Methodol. 2013;13:33. doi: 10.1186/1471-2288-13-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Asselah T., Rubia-Brandt L., Marcellin P., Negro F. Steatosis in chronic hepatitis C: why does it really matter. Gut. 2006;55:123–130. doi: 10.1136/gut.2005.069757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mihm S., Fayyazi, Hartmann H., Ramadori G. Analysis of histopathological manifestations of chronic hepatitis C virus infection with respect to virus genotype. Hepatology. 1997;25:735–739. doi: 10.1002/hep.510250340. [DOI] [PubMed] [Google Scholar]
- 34.Altman D.G., Royston P. The cost of dichotomising continuous variables. BMJ. 2006;332:1080. doi: 10.1136/bmj.332.7549.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Innes H., Goldberg D., Dusheiko G., Hayes P., Mills P.R., Dillon J.F., et al. Patient-important benefits of clearing the hepatitis C virus through treatment: a simulation model. J Hepatol. 2014;60:1118–1126. doi: 10.1016/j.jhep.2014.01.020. [DOI] [PubMed] [Google Scholar]
- 36.Pons M., Rodriguez-Tajes S., Esteban J.I., Marino Z., Vargas V., Lens S., et al. Non-invasive prediction of liver-related events in patients with HCV-associated compensated advanced chronic liver disease after oral antivirals. J Hepatol. 2020;72:472–480. doi: 10.1016/j.jhep.2019.10.005. [DOI] [PubMed] [Google Scholar]
- 37.Audureau E., Carrat F., Layese R., Cagnot C., Asselah T., Guyader D., et al. Personalised surveillance for hepatocellular carcinoma in cirrhosis – using machine learning adapted to HCV status. J Hepatol. 2020;73:1434–1445. doi: 10.1016/j.jhep.2020.05.052. [DOI] [PubMed] [Google Scholar]
- 38.Lopez S.A., Manzano M.L., Gea F., Gutierrez M.L., Ahumada A.M., Devesa M.J., et al. A model based on noninvasvie markers predicts very low hepatocellular carcinoma risk after viral response in hepatitis C virus-advanced fibrosis. Hepatology. 2020;72:1924–1934. doi: 10.1002/hep.31588. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Scottish data used in this study are not publicly available, but can be acquired through successful application to the Public Benefit and Privacy Panel for Health and Social Care (www.informationgovernance.scot.nhs.uk/pbpphsc/home/for-applicants/). The STOP-HCV consortium welcomes collaboration with interested parties. Anonymised samples and clinical data held on the study database are accessible through application to the HCVRUK Tissue Data Access Committee. Contact Professor Indra Neil Guha (neil.guha@nottingham.ac.uk) or Professor William Irving (will.irving@nottingham.ac.uk) for more information. Please note that we cannot share linked NHS digital data with other research groups, or any data variables derived from linked NHS digital data.