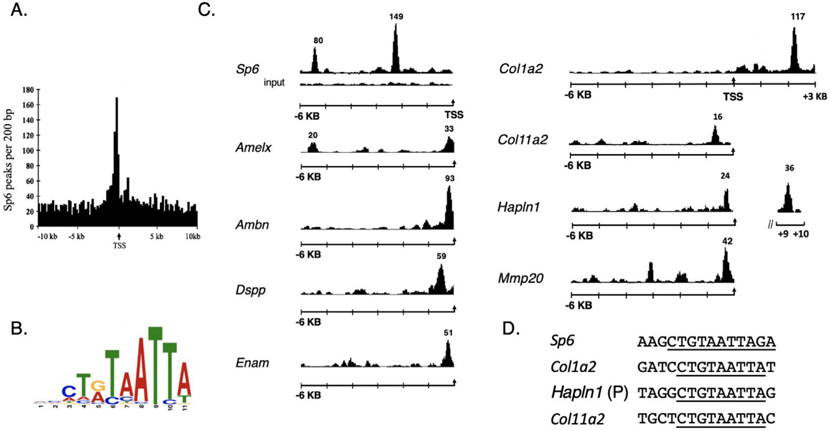
Fig. 1.
ChIP-Seq identifies Sp6 promoter binding sites and a consensus binding motif. In total, 11,436 high confidence Sp6-enriched peaks were identified from tooth mesenchymal tissue. A. 3305 ChIP-seq peaks were identified within 10 Kb of the transcriptional start sites of the target genes. B. MEME bioinformatic analysis of the ChIP-seq data set identified a Sp6 consensus binding motif. C. ChIP-Seq peaks are shown for Sp6 binding sites identified near the transcription start sites (arrow) for the following genes: Sp6, Amelx, Ambn, DSPP, Enam, Col1a2, Col11a2, Hapln1, Mmp20. The black tracing for each gene and the numbers above it represents the Sp6-enriched peak. For this figure, the Sp6 and Col1a2 enriched peak heights are not drawn to scale. D. Extracted peaks from MEME analysis shows CTGTAATTA at SP6, Hapln1, Col11a2, (Col1a2, a major tooth matrix protein did not occur in the Pendleton dataset was evaluated visually).