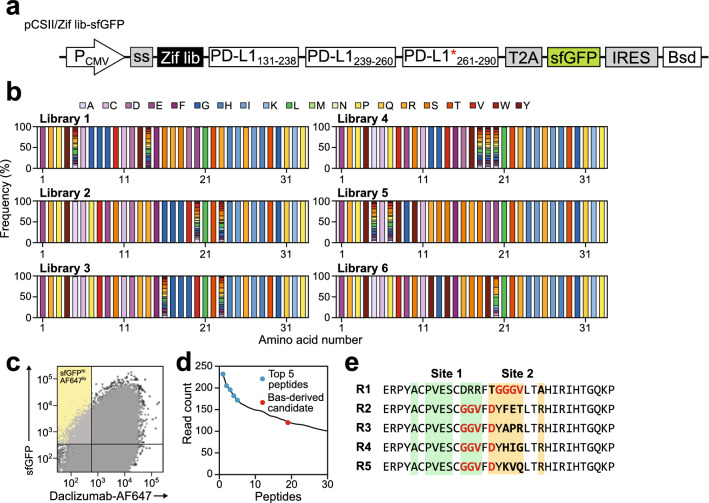
Figure 4.
Antibody-guided screening for Zif-FLAP candidates. (a) Diagram of the Zif-FLAP candidate library construct. Zif lib, Zif-FLAP candidate peptide library. (b) Amino acid frequency in each library set prior to screening. (c) Isolation of the high sfGFP, low AF647 fraction after flow cytometric analysis. K562/CD25 cells transduced with the library construct were incubated with daclizumab-AF647 and then analyzed by flow cytometry for sfGFP expression and AF647 signals. The quadrant in yellow represents the fraction that was sorted out for sequencing. (d) Ranking of peptides in accordance with the read count after NGS analysis. The five most frequently detected sequences are shown by blue circles. The basiliximab (Bas)-derived sequence is indicated by a red circle. (e) Sequences of the top five peptides with the highest read counts. Residues that had been mutated are shown in bold with residues in black representing those that had been fully randomized and those in red representing the CD25-binding residues identified from daclizumab.