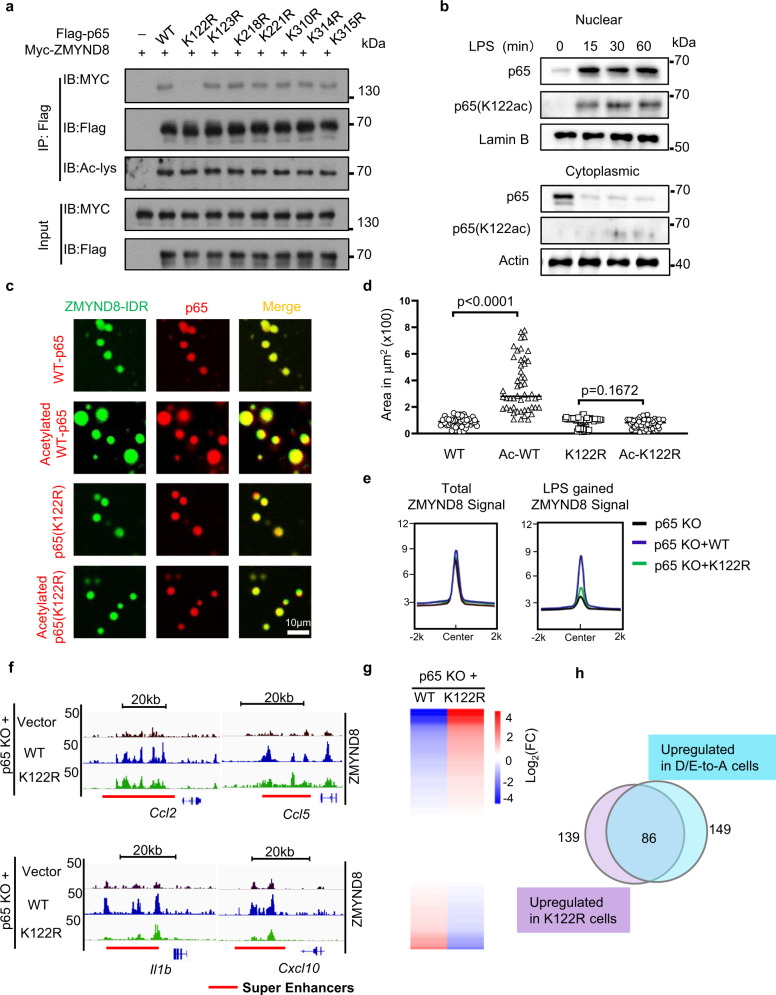
Fig. 6. LPS-induced p65 acetylation at the K122 residue directs the docking of ZMYND8 liquid condensates onto NF-κB associated SEs.
a Co-IP assay between Myc-tagged ZMYND8 and various p65 acetylation site mutants in 293 T cells followed by western blot. Experiments were repeated three times. b The endogenous K122 acetylation level in the cytosol and nucleus of BMDMs was evaluated by western blot at the indicated time after LPS treatment. Experiments were repeated three times. c In vitro droplet formation by incubating GFP-ZMYND8-IDR with mCherry-p65, mCherry-p65-acetylated, mCherry-p65 (K122R), and mCherry-p65 (K122R)-acetylated respectively. Representative images were shown. d Statistical analysis of droplet size formed by GFP-ZMYND8-IDR with mCherry-p65, mCherry-p65-acetylated, mCherry-p65 (K122R), and mCherry-p65 (K122R)-acetylated respectively (n = 50, examined over three independent experiments). Two-tailed Student’s t-test determined p values. e Genome-wide ChIP-Seq analysis of ZMYND8-binding patterns after LPS treatment in different Raw264.7 cells, including p65 KO cell transduced with Vector (p65 KO), p65 KO transduced with WT p65 (p65 KO + WT p65), and p65 KO transduced with K122R mutant (p65 KO + K122R). The average signal intensity of total ZMYND8 peaks (left) and LPS-gained ZMYND8 peaks (right) was shown separately. f Genome browser tracks show that LPS-gained ZMYND8 peaks were recruited to the indicated SE regions in the p65 KO, p65 KO + p65 WT, and p65 KO + K122R Raw264.7 cells after LPS treatment. g Heatmap analysis of RNA-Seq results represent differentially expressed genes regulated by ZMYND8 enriched, latent SEs (from Fig. 2d) in p65 KO + WT p65, p65 KO + K122R Raw264.7 cells after LPS treatment. h The latent SEs regulated genes enhanced in K122R cells are overlapped with latent SE genes upregulated in D/E-to-A cells. Source data are provided as a Source Data file.