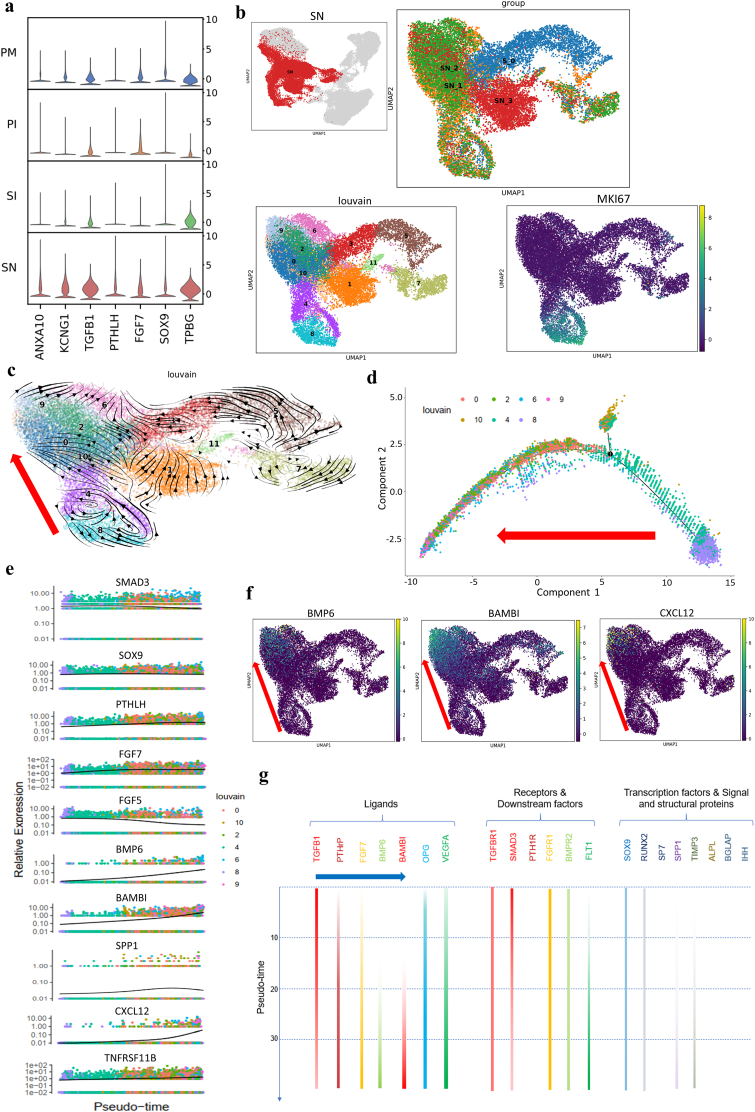
Fig. 3.
Osteogenic process of MSCs cultured on HA scaffolds. a, Stacked violin visualization of the indicated genes highly expressed during HA scaffold-mediated osteogenesis. b, UMAP visualization of subclusters at different time points and MKI67 expression in the SN group. Clusters 8 and 4 represent proliferative cells in a relatively primitive state (S_0, SN at week 0; SN_1, SN at week 1; SN_2, SN at week 2; SN_3, SN at week 3). c, Velocity visualization of subclusters showing the cell development trajectory in the SN group and the direction of osteogenic development (red arrow, clusters 8, 4, 10, 0, 2, 9, 6). d, Monocle pseudotime trajectory showing the progression of osteogenic-lineage clusters (clusters 0, 2, 4, 6, 8, 9, and 10 according to velocity analysis and monocle pseudotime analysis; in Supplementary Fig. 3a, the red arrow shows the cell evolution direction). e, Pseudotime kinetics of the indicated genes during the development of osteogenic-lineage clusters. SOX9 and TNFRSF11B (OPG) showed steady expression levels; FGF5 and SMAD3 showed descending expression tendencies; FGF7 and PTHLH showed increasing expression levels and reached plateaus at the early stage; BMP6, BAMBI, SPP1 and CXCL12 showed elevated expression at the late stage. f, Expression distribution in UMAP visualization of the indicated genes (the red arrow shows the osteogenic direction). g, Summary of key signaling molecules involved in osteogenesis in the SN group showing the chronological activation of osteogenic signaling pathways (blue arrow). The expression level is indicated by the color intensity. The expression levels of genes not included in the pseudotime kinetics are supplied in Supplementary Fig. 3b because of low expression differences, and expression-deficiency strips show a lack of expression of PTH1R, SP7, ALPL, BGLAP and IHH.