Abstract
hnRNPK is a multifunctional protein that plays an important role in cancer cell proliferation and metastasis via its RNA- and DNA-binding properties. Previously we showed that cell-penetrating peptides derived from the RGG RNA-binding domain of SAFA (hnRNPU) disrupt cancer cell proliferation and survival. Here we explore the efficacy of a peptide derived from the RGG domain of hnRNPK. This peptide acts in a dominant-negative manner on several hnRNPK functions to induce death of multiple types of cancer cells. The peptide phenocopies the effect of hnRNPK knockdown on its mRNA-stability targets such as KLF4 and EGR1 and alters the levels and locations of long non-coding RNAs (lncRNAs) and proteins required for nuclear and paraspeckle formation and function. The RGG-derived peptide also decreases euchromatin as evidenced by loss of active marks and polymerase II occupancy. Our findings reveal the potential therapeutic utility of the hnRNPK RGG-derived peptide in a range of cancers.
Keywords: hnRNPK, cancer therapy, lncRNA, nuclear speckles, cell-penetrating peptides, dominant-negative, nuclear paraspeckles, chromatin marks, RNA processing
Graphical abstract

Here we show that a peptide derived from the RGG domain of hnRNPK acts in a dominant-negative manner on several hnRNPK functions to induce death of multiple types of cancer cells without harming normal cells. Our findings reveal the potential therapeutic utility of the hnRNPK RGG-derived peptide in a range of cancers.
Introduction
Heterogeneous nuclear ribonucleoprotein K (hnRNPK) is a member of the hnRNP family of RNA-binding proteins (RBPs) with numerous and complex molecular functions. It has critical roles in diverse cellular processes including proliferation, DNA repair, differentiation, and cancer progression.1, 2, 3, 4 In addition to regulating mRNA transport, splicing, stability, and translation, it mediates interactions between nucleic acids, long non-coding RNAs (lncRNAs), and partner proteins to activate or repress transcription networks downstream of a variety of signaling pathways such as the p53, MYC, and FOS networks. hnRNPK plays a central role in the DNA damage response and apoptosis in coordination with p53. The formation and function of nuclear paraspeckles and recruitment of splicing factors to nuclear speckles and active chromatin all require hnRNPK.
Recent chromatin immunoprecipitation sequencing (ChIP-seq) experiments revealed that chromatin occupancy by hnRNPK positively correlates with active chromatin marks, DNase I hypersensitive sites, polymerase II (Pol II) binding, and active transcription.5 Repressive chromatin marks are depleted from hnRNPK-bound promoters, and knockdown of hnRNPK leads to Pol II dissociation from many HNRNPK bound regions.6 hnRNPK interacts with the TATA-binding protein (TBP) of the RNA polymerase machinery.4,7 This protein plays a critical role in higher-order chromatin structure and remodeling as a matrix attachment region binding protein to stabilize chromatin loops.8 It also functions in transcriptional repression by interacting with Polycomb complexes9 and promotes transcriptional termination through the XRN2 pathway.5
In addition to DNA-dependent molecular functions, hnRNPK regulates many aspects of mRNA, lncRNA, and microRNA (miRNA) processing, stability, and nuclear retention.10 It binds nascent mRNAs and recruits RBPs and splicing machinery to nuclear speckles and to chromatin of actively spliced genes.10,11 hnRNPK binds to GRHL3, KLF4, and ZNF750 transcripts and decreases their mRNA stability to prevent premature differentiation and regulate apoptosis in epidermal stem cells.6 By binding to the 3′ UTR of CDKN1A, hnRNPK negatively regulates its translation in a neuronal context.12 In contrast, it can directly bind to the 5′ UTR of the MYC transcript to promote ribosomal engagement.13 hnRNPK plays a role in processing, nuclear retention, and function of lncRNAs in numerous nuclear subdomains, including nuclear speckles and paraspeckles.10 Disruption of a short interspersed nuclear element (SINE) in MALAT1 lncRNA disrupts its interaction with hnRNPK, leading to increased cytoplasmic MALAT1, abnormal nuclear speckle morphology and function, and DNA damage and apoptosis.14 hnRNPK is required for alternative 3′ end processing of NEAT1 to generate the isoform required for nuclear paraspeckle formation, maintenance, and function.15, 16, 17, 18
Depending on context, hnRNPK can function as an oncogene or tumor suppressor.19, 20, 21, 22 Its overexpression in several tumors (breast, hepatocellular, colorectal, melanoma) correlates with poor outcomes and advanced disease,23, 24, 25, 26, 27 and depletion of hnRNPK reduces proliferation of pancreatic and renal cancer lines.28,29 In other contexts, its functions as a tumor suppressor are clear: it is a p53 coactivator30,31 and is actuated downstream of the ATR/ATM pathways during the DNA-damage response.32,33 Hnrnpk+/− mice have decreased survival due to myeloproliferation, lymphomas, and hepatocellular carcinomas.34
Cell-penetrating peptides (CPPs) are composed of short stretches of amino acids that facilitate translocation of cargo molecules across cell membranes.35 This strategy has been employed to deliver dominant-negative peptides that abrogate the function of the oncoproteins MYC and ATF5 and are now in clinical trials.36,37 In addition to these targets, numerous peptides have been developed with effects on gastric and colon cancers,38 breast cancer,39 glioma,40 and skin cancer.41 We recently developed novel RBM39- and hnRNPU-derived dominant-negative CPPs that reduce the survival and proliferation of a range of cancer cells.42,43 We have demonstrated that RBM39-derived CPPs disrupt its interaction with the MLL1 complex and its DNA targets, while a peptide derived from the RGG domain of SAFA (also known as hnRNPU) disrupts its interaction with target RNAs. In both cases, the resulting effects on the epigenetic landscape, transcription, and splicing decrease the growth and survival of multiple types of cancer cells while leaving normal cells unharmed. hnRNPK harbors three K homology (KH) domains that are highly conserved across other poly(C)-binding proteins and facilitate binding to poly(C) tracts in both RNA and single-stranded DNA.44 It also contains a K-protein-interaction (KI) domain harboring an RGG box that facilitates protein-protein interactions and nucleic acid binding.8,22,45,46 Because the RGG RNA-binding domains of SAFA and hnRNPK are highly related, here we evaluate the effects of a peptide derived from the RNA-binding domain of hnRNPK on the proliferation, survival, and transcriptional and epigenetic features of various cancer cells.
Results
Cell-penetrating peptide derived from the RGG domain of hnRNPK decreases survival of multiple types of cancer cells
We hypothesized that the RGG domain of hnRNPK would have dominant-negative activities and phenocopy at least some aspects of hnRNPK loss of function.28,29,47,48 To test this, we synthesized a peptide containing amino acids 241–270 of the hnRNPK RGG-domain conjugated to Penetratin CPP49 and to a His6-tag (Figure 1A, CPP-hnRNPK-RGG); His-tagged Penetratin served as negative control (CPP-Neg). Initially, we evaluated their cellular penetration by treating HEK293 cells with 10 μM peptide in medium for 6 h and used anti-His antibody to detect the peptide by immunofluorescence. Both peptides exhibited uniform penetration into the cytoplasm and nucleus (Figures S1A and S1B). Next, we treated cancer cell lines (MDA-MB231, triple-negative breast cancer; DU145, prostate cancer; HT1080, fibrosarcoma; UMUC3, bladder cancer) with peptides at 10 μM for 24 h. Crystal violet staining showed that CPP-hnRNPK-RGG reduced the total number of all four cell types (Figure 1B, quantitated in 1C). More detailed analysis showed that this decrease was due to decreased viability (Figure 1D). In contrast, treatment of MCF10A cells, a transformed but non-malignant breast cancer cell line, or primary human foreskin fibroblasts (HFFs) had no effect on their proliferation or survival (Figures 2A–2C). The HCT116 colon cancer cell line was also minimally affected (Figure 2). Thus CPP-hnRNPK-RGG has cell-type-specific functions.
Figure 1.
CPP-hnRNPK-RGG peptide decreases viability of MDA-MB231, DU145, HT1080, and UMUC3 cancer cells
(A) Amino acid sequences of CPP-Neg (negative control CPP) and CPP-hnRNPK-RGG (CPP conjugated with hnRNPK-derived RGG domain). The Penetratin sequence is in red and His-tag is in blue. (B) Representative light-microscopic images of MDA-MB231, DU145, HT1080, and UMUC3 cancer cells stained with crystal violet. Cells were treated for 24 h with 10 μM concentration of the synthetic peptide indicated at the side. Scale bars, 50 μm. (C) Quantification of total cell number of all four cancer cell types measured with a hemocytometer after 24 h of treatment with 10 μM CPP-Neg or CPP-hnRNPK-RGG. (D) Quantification of viability (absorbance, 490 nm, y axis) as measured with the CellTiter-Blue viability assay colorimetric method after 24 h of treatment with 10 μM CPP-Neg or CPP-hnRNPK-RGG. ∗p < 0.05, ∗∗p < 0.01.
Figure 2.
Effects of CPP-hnRNPK-RGG are cell-type specific
(A) Representative light-microscopic images of crystal violet-stained MDA-MB231 breast cancer cells, the benign MCF10A breast cells, primary human fibroblasts (HFF), and HCT116 colon cancer cells. Cells were treated for 24 h with 10 μM peptide indicated at the left. Scale bars, 50 μm. (B) Quantification of total cell number of all four cell types measured with a hemocytometer after 24 h of treatment with 10 μM CPP-Neg or CPP-hnRNPK-RGG. (C) Quantification of cell viability (absorbance, 490 nm, y axis) as in Figure 1. ∗p < 0.05, ∗∗p < 0.01.
Peptides derived from the RGG domains of hnRNPA1 and hnRNPF have no effect on cancer cell proliferation or viability
We synthesized additional CPPs from the RNA-binding domains of the related proteins hnRNPA1 (amino acids 218–240, CPP-hnRNPA1-RGG) and hnRNPF (amino acids 176–186, CPP-hnRNPF-RBD) (Figure S2A) and confirmed that these peptides penetrate HEK293 cells (Figures S1C and S1D). We then employed the same assays to evaluate cell number and viability but observed no significant effect of these peptides on any of the cell lines (Figures S2B–S2D). These findings indicate that there are specific attributes of the peptide derived from the RNA-binding domain of hnRNPK that confer dominant-negative activity.46,50, 51, 52
Cancer cell apoptosis/necrosis in response to CPP-hnRNPK-RGG is dosage dependent
We used trypan blue to measure the number of dead cells in vehicle-treated, CPP-Neg, and CPP-hnRNPK-RGG cells (Figure 3A). While the numbers of dead cells were minimally different between vehicle and CPP-Neg treatments, CPP-hnRNPK-RGG treatment markedly increased the number of trypan blue-stained (dead) cells. We then used an Annexin V-based assay to examine the apoptosis/necrosis response to varying doses of CPP-hnRNPK-RGG (Figure 3B). Equal numbers of each cell type were treated with 1, 5, and 10 μM peptide and assayed at regular intervals over 24 h. When treated with 5 μM peptide, all cell types had a modest increase in apoptosis/necrosis relative to treatment with either 1 μM CPP-hnRNPK-RGG or 1 μM CPP-Neg. In contrast, 10 μM treatment revealed differential sensitivity of the cancer cell lines: MDA-MB231 cells were the most sensitive, with a maximal effect at 8 h, while HT1080 and DU145 cells were approximately 40% less sensitive, and their effect peaked at 4 h; UMUC3 cells had a continuous increase in the number of apoptotic cells over the 24-h monitoring period.
Figure 3.
Cell death in response to CPP-hnRNPK-RGG treatment is dosage sensitive and cell-type specific
(A) Quantification of trypan blue-positive dead cells. (B) Quantification of apoptosis/necrosis as measured in fluorescence units detected with the RealTime-Glo Annexin V apoptosis and necrosis assay in response to 1, 5, and 10 μM peptide treatments assayed at the times shown below each graph. Blue and orange lines represent CPP-Neg and CPP-hnRNPK-RGG peptide treatments, respectively. ∗p < 0.05, ∗∗p < 0.01.
CPP-hnRNPK-RGG does not alter cancer cell-cycle kinetics
The differential sensitivity of these cancer cell types to CPP-hnRNPK-RGG peptide treatment suggests divergent mechanisms underlying their response. Previously, we showed that treatment of these cancer lines with hnRNPU-RGG-derived peptide resulted in decreased expression of E2F1-responsive genes and perturbed the cell cycle in a cell-type-specific manner.42 Others have shown that hnRNPK loss of function leads to reduced expression of cell-cycle genes, blocking the cell cycle at different stages depending on cell type.53,54 In the case of CPP-hnRNPK-RGG-treated cells, flow cytometry analysis of propidium iodide-stained cells showed no alteration in cell-cycle distribution relative to CPP-Neg, with the exception of a slight increase in HT1080 cells in S phase (Figure S3A). Although CPP-hnRNPK-RGG treatment caused decreases in E2F1 and CDC2 transcript levels, their downstream transcripts were largely unaffected (Figure S3B), a finding that has been reported elsewhere and attributed to redundancy of E2F proteins.55 These cell-cycle results are congruent with the findings in Figures 1 and 3, showing that the mechanism of decreased cell number in response to CPP-hnRNPK-RGG is induction of cell death.
CPP-hnRNPK-RGG increases transcript levels of proapoptotic mRNAs normally destabilized by hnRNPK
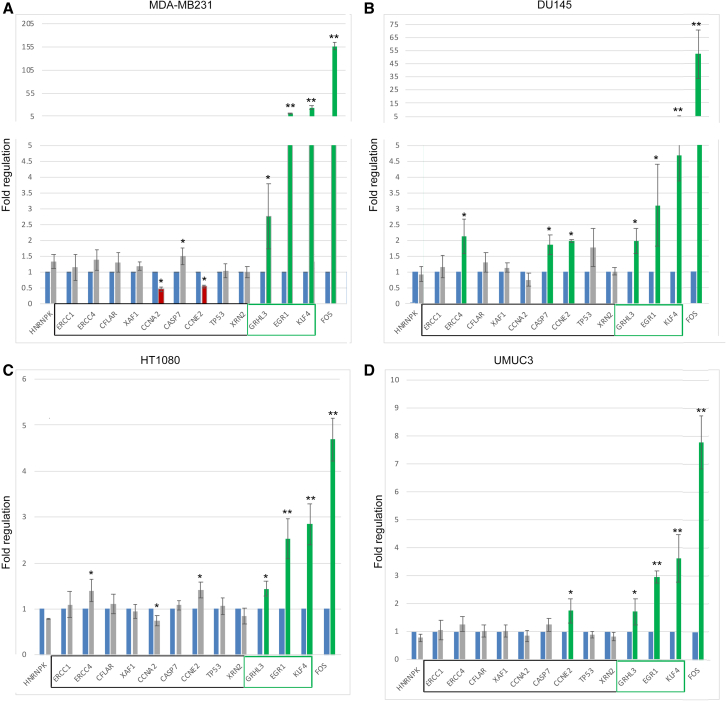
hnRNPK regulates the levels of its target transcripts by both co- and post-transcriptional mechanisms.44 We tested whether hnRNPK-RGG alters the levels of transcripts regulated by both mechanisms. The levels of several known transcriptionally regulated targets5,6,25,48,56 were not or only modestly affected by CPP-hnRNPK-RGG (black boxed gene names in Figure 4). Transcripts previously reported to be regulated by hnRNPK-mediated degradation in some cellular contexts were markedly increased in all four cell types (GRHL3, EGR1, KLF4; green boxed in Figure 4), as was seen with hnRNPK knockdown.5,6 FOS transcripts were markedly increased in all four cell types.
Figure 4.
CPP-hnRNPK-RGG dysregulates the abundance of known hnRNPK target mRNAs
(A–D) qRT-PCR analysis of mRNA transcripts after 24 h of 10 μM peptide treatment. Cell types are labeled: (A) MDA-MB231, (B) DU145, (C) HT1080, (D) UMUC3. Green and red bars indicate increased and decreased transcript levels, respectively. Error bars show standard deviation. In other studies,5,6,46 the transcripts named in black boxes at the bottom of the graphs have been shown to be regulated by hnRNPK at the transcriptional level and those in green boxes at the level of mRNA stability. ∗p < 0.05, ∗∗p < 0.01.
CPP-hnRNP-RGG disrupts amount and location of nuclear speckle- and paraspeckle-associated lncRNAs and RNA-binding proteins
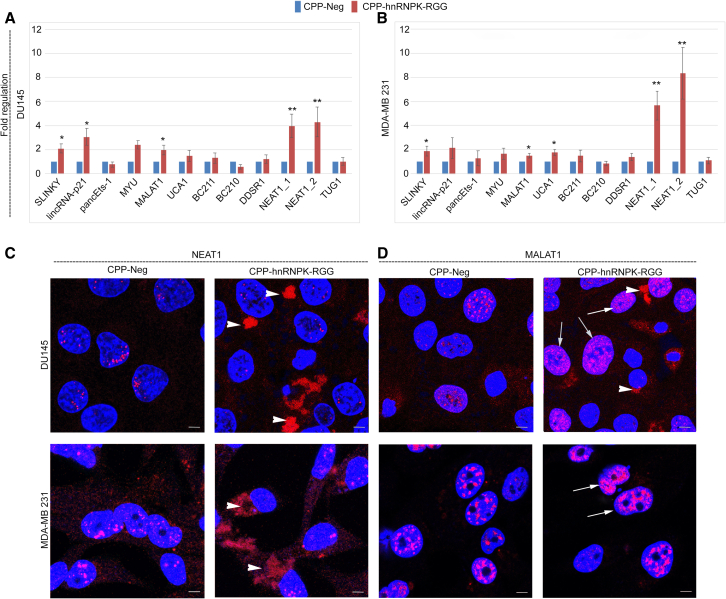
The contribution of hnRNPK to post-transcriptional regulation of gene expression occurs in part via its roles in nuclear retention of lncRNAs and mRNAs and its functions in nuclear paraspeckles and speckles.11,57 It plays crucial roles in 3′ end processing and nuclear retention of the paraspeckle transcript NEAT1, and in recruitment and retention of MALAT1 to nuclear speckles. These hnRNPK/lncRNA interactions (and others) are critical to many cellular processes, including the formation and function of these nuclear domains.10 We assayed the levels of several cancer-related, hnRNPK-interacting lncRNAs58 in DU145 and MDA-MB231 cells (Figures 5A and 5B), which revealed elevated levels of many of these, the most marked being increased levels of NEAT1_1 and NEAT1_2 relative to CPP-Neg. Further, 10 μM CPP-hnRNPK-RGG decreased the ratio of NEAT1_1 to NEAT1_2 (Figure S4), suggesting that the RGG domain of hnRNPK is not required for alternative 3′ end processing of NEAT1_1 to generate NEAT1_2. Evaluation of NEAT1 transcripts by fluorescence in situ hybridization (FISH) showed that CPP-hnRNPK-RGG decreased the number of paraspeckles in both cell types, and this was associated with mislocalization of NEAT1 transcripts to the cytoplasm (Figures 5C, arrowheads, and S5A). CPP-hnRNPK-RGG treatment altered the pattern and intensity of signal of the nuclear speckle lncRNA MALAT1, resulting in a dense network of fine nuclear bodies (Figure 5D, arrows) and in transcripts also mislocalized to the cytoplasm in DU145 cells (Figures 5D, arrowheads, and S5B). The changes we observed in NEAT1 and MALAT1 staining are quantitated in Figure S6.
Figure 5.
Altered levels and location of lncRNAs, nuclear speckles, and paraspeckles induced by CPP-hnRNPK-RGG
(A and B) qRT-PCR analysis of levels of lncRNA transcripts listed at bottom in (A) DU145 and (B) MDA-MB231 cells in response to treatment with control or CPP-hnRNPK-RGG peptides at 10 μM for 24 h. (C) Fluorescence in situ hybridization to detect NEAT1 lncRNA in DU145 and MDA-MB231 cells. CPP-hnRNPK-RGG at 10 μM for 24 h decreases the number of nuclear paraspeckles (punctate nuclear signal) while increasing the presence of transcripts in the cytoplasm (arrowheads) in comparison to control peptide. Scale bars, 10 μm. Additional images are presented in Figure S5. (D) Fluorescence in situ hybridization to detect MALAT1 in DU145 and MDA-MB231 cells. CPP-hnRNPK-RGG at 10 μM for 24 h increases the number of cells with diffuse, bright staining (arrows) and cytoplasmic staining (arrowheads). Scale bars, 10 μm. Quantitative FISH results are presented in Figure S6. ∗p < 0.05, ∗∗p < 0.01.
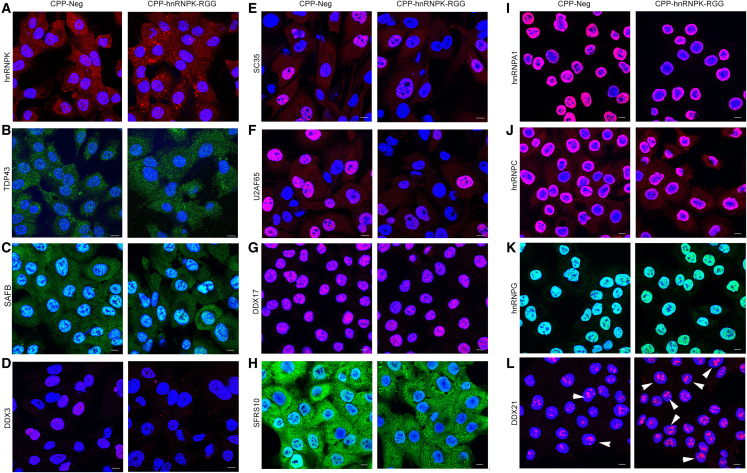
Abnormal function of hnRNPK and abnormal number or composition of nuclear speckles and paraspeckles would be predicted to affect the localization of RBPs and splicing factors. For example, MALAT1 binds splicing factors in nuclear speckles and regulates their distribution and function. We found that treatment with 10 μM CPP-hnRNPK-RGG increased the number of cells with dense cytoplasmic aggregates of hnRNPK (Figure 6A), but had minimal effect on TDP43 or SAFB (Figures 6B and 6C). CPP-hnRNPK-RGG markedly altered the level and/or location of several RBPs in DU145 cells (Figure 6): DDX3, SC35, U2AF65, DDX17, and SFRS10 are splicing factors that are decreased in the nucleus, and SFRS10 is also decreased in the cytoplasm (Figures 6D–6H). hnRNPA1 and hnRNPC are both nuclear envelope proteins that also have splicing functions: hnRNPA1 activates splicing, while hnRNPC competes with U2AF65 to repress splicing of shared target RNAs.59 Treatment with10 μM CPP-hnRNPK-RGG decreased hnRNPA1, while hnRNPC was increased (Figures 6I and 6J). The levels of hnRNPG were not significantly changed (Figure 6K). Increased size and number of nucleoli is a stress response and part of the apoptotic process,60 and we detected an increase in the number of cells with large, dense DDX21-positive nucleoli with CPP-hnRNPK-RGG treatment (Figure 6L, arrowheads). These findings are quantitated in Figure S7. Similar findings in the nuclear signal of the splicing factors SC35, U2AF65, and SFSR10 and hnRNPs A1, C, and G were seen in MDA-MB231 cells (Figure S8).
Figure 6.
Altered levels and localization of splicing factors and RNA-binding proteins in response to CPP-hnRNPK-RGG in DU145 cells
(A–L) Fluorescence immunocytochemical detection of the indicated proteins in DU145 cells after 24 h of treatment with 10 μM peptides. (A) hnRNPK, (B) TDP43, (C) SAFB, (D) DDX3, (E) SC35, (F) U2AF65, (G) DDX17, (H) SFRS10, (I) hnRNPA1, (J) hnRNPC, (K) hnRNPG, (L) DDX21. White arrowheads highlight cells with enlarged, dense DDX21-positive nuclei. Scale bars, 10 μm. Quantitation of these findings is presented in Figure S7.
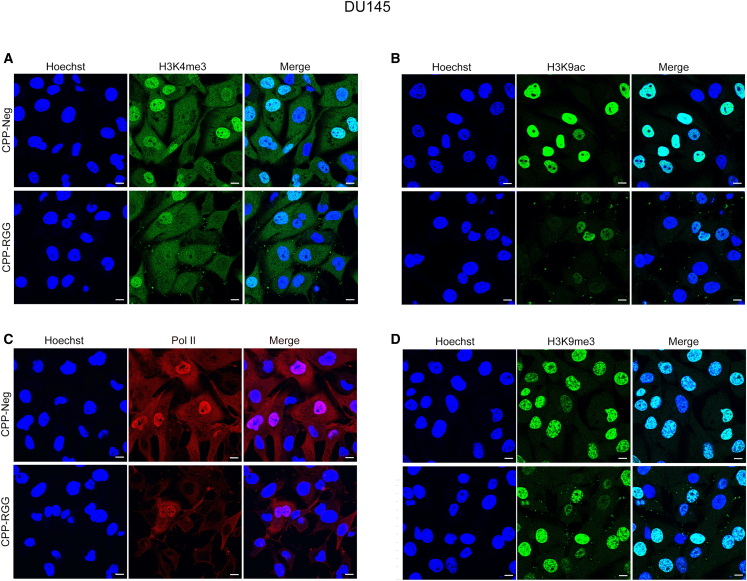
CPP-hnRNP-RGG reduces global active epigenetic marks in cancer cells
Previous reports demonstrated that hnRNPK can regulate transcription of its targets by directly interacting with their promoters and influencing histone marks.4,61 Thus, we tested if CPP-hnRNPK-RGG alters Pol II occupancy or bulk levels of the active marks H3K4me3 and H3K9ac, or the H3K27me3 repressive mark. Immunofluorescence assays show that CPP-hnRNPK-RGG treatment of DU145 cells at 10 μM results in markedly decreased levels of the active marks H3K4me3 and H3K9ac, as well as Pol II occupancy, with minimal effect on the repressive mark H3K9me3 (Figures 7A–7D, quantitation in Figure S9). Immunoblotting of DU145, HT1080, UMUC3, and MDA-MB231 cell lysates also showed decreased levels of active marks H3K4me3 and H3K9ac, with no effect on the H3K27me3 repressive mark (Figure S10). Together, these results strengthen the correlation between active chromatin and hnRNPK and suggest that this epigenetic function is mediated by the RGG domain.
Figure 7.
Active chromatin marks and Pol II occupancy are decreased by CPP-hnRNPK-RGG in DU145 cells
Fluorescence immunocytochemical detection of histone marks and Pol II in DU145 cells after 24 h of 10 μM peptide treatment. (A) H3K4me3, (B) H3K9ac, (C) Pol II, and (D) H3K9me3. Scale bars, 10 μm. Quantitative results are presented in Figure S9.
Discussion
hnRNPK has central roles in regulating gene expression at multiple levels and in multiple cellular compartments, including transcription via direct promoter binding; Pol II binding and establishing and/or maintaining active chromatin structure; alternative mRNA splicing, 3′ end processing, and miRNA biogenesis; nuclear retention and stability of mRNAs and lncRNAs; and function and location of partner RBPs. Our results demonstrate that treatment with CPP-hnRNPK-RGG peptide disrupts many of these functions, resulting in cellular stress and apoptosis in MDA-MB231, DU145, HT1080, and UMUC3 cancer cells. These effects were not seen in HCT116 hepatocarcinoma cells, primary human fibroblasts, or the benign MCF10A breast cell line, suggesting that there are cell-type-specific differences in the function of the hnRNPK-RGG domain. The cell-type-specific variations we observe in response to CPP-hnRNPK-RGG are consistent with gene- and cell-type-specific transcriptional activation and repression activities by hnRNPK reported previously.4,62, 63, 64, 65
Elevation of KLF4 and EGR1, both of which are well-established drivers of apoptosis,66,67 by CPP-hnRNPK-RGG may occur via disruption of the binding and/or processing functions of hnRNPK on these transcripts, as has been reported elsewhere.5,6 The mechanism of peptide-mediated increases in FOS transcripts that we observe is unclear, but this finding has been reported in cells undergoing apoptosis.68 Removal of the RGG domain of hnRNPK was shown to disrupt its binding to an RNA probe containing 32 nucleotides of the c-FOS mRNA, and hnRNPK regulation of the FOS promoter in transactivation assays is thought to occur via hnRNPK binding of the nascent mRNA.69
The functions of nuclear paraspeckles are yet to be completely elucidated, but they are an established domain for mRNA adenosine-to-inosine (A-I) editing and retention. Paraspeckles are highly sensitive to a variety of cellular stresses that alter their number, distribution, and ability to release or retain transcripts via effects on nuclear export. Alterations in miRNA biogenesis in paraspeckles in response to stressors is another mechanism whereby this compartment regulates gene expression. hnRNPK controls alternative 3′ end processing of NEAT1 to generate the isoform required for nuclear paraspeckle formation, maintenance, and function, and we found that CPP-hnRNPK-RGG causes an increase in transcript levels of both isoforms of NEAT1, which manifests as increased transcripts in the cytoplasm, while the number of paraspeckles is decreased in both DU145 and MDA-MB231 cells. These observations suggest that the RGG domain of hnRNPK facilitates retention of NEAT1 in paraspeckles.
Nuclear speckles (NS) are known to house splicing and other RNA-processing machinery and, more recently, their role in “boosting” active transcription and Pol II occupancy has been revealed as a mechanism that coordinates multiple aspects of expression regulation.70 The lncRNA MALAT1 is critical to this aspect of nuclear speckle function by recruiting transcriptional machinery to active chromatin and interacting with chromatin at actively spliced genes.71 Discovery of a consensus sequence within lncRNA SINEs that is bound by hnRNPK and mediates nuclear accumulation has far-reaching implications,57 as disruption of this element in the MALAT1 lncRNA resulted in increased cytoplasmic MALAT1, abnormal nuclear speckle morphology and function, and DNA damage and apoptosis.14 The hnRNPK functional domain(s) required for retention of MALAT1 is unknown; however, our data show that CPP-hnRNPK-RGG causes increased MALAT1 levels, altered nuclear speckle size and distribution, and mislocalization of this lncRNA to the cytoplasm in DU145 cells, similar to the effects of deleting the 3′ SINE in MALAT1. All of these observations suggest that the RGG domain plays a role in the hnRNPK/MALAT1 SINE interaction. The level of cytoplasmic hnRNPK relative to that in the nucleus also increased in response to CPP-hnRNPK-RGG (Figures 6A and S7A), suggesting a contribution of the RGG domain in nuclear retention of hnRNPK in these cells.46 Additional effects of CPP-hnRNPK-RGG on nuclear speckles are evident in the altered levels and locations of splicing factors and other RBPs.
In addition to effects on active transcription due to alterations in nuclear speckles/MALAT1, CPP-hnRNPK- RGG decreased bulk levels of activating histone marks and levels of nuclear Pol II. Increased levels of lincRNA-p21 may also contribute to the decrease in active chromatin that we observed.72
Collectively, these findings support our hypothesis that the peptide disrupts hnRNPK-mediated recruitment of, or interaction with, proteins and lncRNAs that confer active marks and regulate higher-order chromatin structure. hnRNPK has known roles in these processes and also is required for the binding of Pol II to active sites of transcription.5,6 The observed changes in active chromatin would be expected to have widespread consequences on gene expression that are beyond the scope of the present study. Clearly, additional investigation into the factors that engage with hnRNPK to establish active chromatin in cancer cells is warranted, as is determining the molecular roles of the RGG domain in these events.
Our published work shows that a dominant-negative peptide derived from the RGG domain of SAFA (hnRNPU) has effects on cancer cell survival, histone marks, and other key processes that are similar to those of CPP-hnRNPK-RGG.42,43 Despite the similarity in structure between the RGG domain of hnRNPK and hnRNPU and those of other hnRNPs, peptides derived from the RGG domains of hnRNPA1 and hnRNPF had no effect on survival of the cancer cells we tested. It could be that these hnRNPs have more limited roles in cancer cells and/or that their RGG domains have molecular functions different from those of hnRNPK and hnRNPU. Future studies will be directed at determining the respective interacting partners of hnRNPK and hnRNPU in cancer cells. Unbiased studies are needed to determine and compare the breadth of altered chromatin structure and gene expression in response to CPP-hnRNPK-RGG in different types of cancer cells.
Pharmacologic or nucleic acid-based methods to inhibit key oncogenic factors are the most common strategies to disrupt cancer progression or metastasis. However, many of these interventions are directed at molecules and processes that are also required in normal cells, resulting in significant toxicity. We have shown that dominant-negative peptides derived from the RNA-binding domains of hnRNPU and RBM39 (also known as CAPERα) have minimal effects on normal cells because they disrupt cancer-cell-specific interactions and molecular functions42,43 and may have significant clinical advantages over less targeted approaches. Here, we provide support for further exploration and development of a peptide with dominant-negative functions in hnRNPK, called CPP-hnRNPK-RGG, as a therapeutic peptide.
Materials and methods
Cell culture
MDA-MB231, UMUC3, HCT116, DU145, HFF1, and HT1080 cell lines were obtained from ATCC. These cell lines were maintained in Dulbecco's modified Eagle's medium (cat. no. 30-2002) with 10% fetal bovine serum. The MCF10A cell line was obtained from ATCC and were cultured in MEBM complete medium (kit cat. no. CC-3150 + 100 ng/mL cholera toxin). Cells were maintained in a humid incubator with 5% CO2 at 37°C.
Antibodies
The following antibodies were used in this study: R-IgG (SC-2027), m-IgG (SC-2025), actin (SC-47778), H3K4me3 (Cell Signaling, 9751; active motif 39159), H3K27me3 (Cell Signaling, 9733), H3K27ac (ab4729), H3K9ac (ab176916), rabbit polyclonal Ki67 (Vector Labs), hnRNPK (SC-28380), TDP43 (ab133547), SAFB1 (A300-811A), DDX3 (SC-365768), SC35 (ab11826), U2AF65 (SC-53942), DDX17 (SC-130650), SFRS10 (Bethyl, A305-011A), hnRNPA1 (SC-32301), hnRNPC (SC-32308), hnRNPG (Cell Signaling D7C2V), and DDX21 (SC-376953).
For the immunohistochemistry (IHC) studies, primary antibody was used at a 1:200 dilution and secondary at 1:1,000. For western blots, a 1:1,000 dilution was used for the primary and 1:10,000 for the secondary.
Crystal violet staining
Crystal violet staining was performed as in Puvvula and Moon.42 Briefly, cells were grown in six-well plates in complete growth medium. When they reached 60% confluence, the growth medium was replaced with Opti-MEM medium supplemented with peptide at the concentrations shown in the figure legends. Cells were incubated for the times indicated and fixed with 1% paraformaldehyde (PFA) for 10 min. Subsequently, the cells were stained with 100 μL 0.1% crystal violet solution for 2 h. Crystal-violet-stained cells were observed and recorded using inverted optical microscopy.
Cell count analysis
Cells were plated in six-well dishes and incubated with synthetic peptides in Opti-MEM at the concentrations and times indicated in the figures or legends, and then the cells were trypsinized and counted using a hemocytometer.
RNA isolation and reverse transcription-PCR
Total RNA was prepared as per the manufacturer's protocols. We used the RNeasy Mini Kit (cat. no. 74104) from Qiagen to extract the RNA and then prepared cDNA using the EcoDry Premix Double Primed kit (Clontech). Quantitative RT-PCR was performed using SsoFast EvaGreen Supermix (Bio-Rad) as per the manufacturer's protocol. RT-PCR primer sequences will be provided upon request.
Synthetic peptides
LifeTein synthesized hnRNPK-, hnRNPA1-, and hnRNPF-derived peptides at purity >75%. Peptides were dissolved in PBS at 1 mg/mL and then applied to cells at the concentrations stated in the legends in Opti-MEM medium.
CellTiter-Blue
Cell Viability Assay (cat. no. G8080, Promega) and RealTime-Glo Annexin V Apoptosis and Necrosis Assay (JA1011, Promega) were performed as per Promega's specified protocols.
lncRNA FISH
NEAT1 (SMF-2037-1) and MALAT1 (2035-1) Stellaris FISH probes were purchased from Biosearch Technologies and RNA-FISH was performed as per the manufacturer's protocol. Briefly, cells were grown on 18-mm round coverglasses in 12-well cell culture plates and subjected to peptide treatment for 24 h at 10 μM concentration. At the end of the treatment, cells were washed with 1× PBS and fixed for 10 min in fixation buffer. Cells were permeabilized with 70% ethanol for at least 1 h in the cold. Before hybridization, cells were pretreated once with wash buffer A. Cells were incubated with 100 μL of hybridization buffer containing 125 nM lncRNA probes overnight. Cells were washed with wash buffers A and B and stained with Hoechst at the end of the procedure. LncRNA signals were visualized and recorded by confocal microscopy.
Data availability
All data are contained in the article and supplemental data files.
Acknowledgments
We thank Diana Lim and the University of Utah Molecular Medicine Program for creating the graphical abstract. This study was funded by the Geisinger Clinic/Weis Center for Research.
Author contributions
P.K.P. designed and performed experiments, co-wrote the manuscript, and prepared figures. S.B. performed experiments and proofread the manuscript. A.M. analyzed the experiments, co-wrote the manuscript, and prepared figures.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omto.2021.10.004.
Contributor Information
Pavan Kumar Puvvula, Email: pkpuvvula@geisinger.edu.
Anne M. Moon, Email: ammoon@geisinger.edu.
Supplemental information
References
- 1.Krecic A.M., Swanson M.S. hnRNP complexes: composition, structure, and function. Curr. Opin. Cell Biol. 1999;11:363–371. doi: 10.1016/S0955-0674(99)80051-9. [DOI] [PubMed] [Google Scholar]
- 2.Bomsztyk K., Van Seuningen I., Suzuki H., Denisenko O., Ostrowski J. Diverse molecular interactions of the hnRNP K protein. FEBS Lett. 1997;403:113–115. doi: 10.1016/s0014-5793(97)00041-0. [DOI] [PubMed] [Google Scholar]
- 3.Tomonaga T., Levens D. Activating transcription from single stranded DNA. Proc. Natl. Acad. Sci. U S A. 1996;93:5830–5835. doi: 10.1073/pnas.93.12.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michelotti E.F., Michelotti G.A., Aronsohn A.I., Levens D. Heterogeneous nuclear ribonucleoprotein K is a transcription factor. Mol. Cell Biol. 1996;16:2350–2360. doi: 10.1128/mcb.16.5.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mikula M., Bomsztyk K., Goryca K., Chojnowski K., Ostrowski J. Heterogeneous nuclear ribonucleoprotein (HnRNP) K genome-wide binding survey reveals its role in regulating 3’-end RNA processing and transcription termination at the early growth response 1 (EGR1) gene through XRN2 exonuclease. J. Biol. Chem. 2013;288:24788–24798. doi: 10.1074/jbc.M113.496679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li J., Chen Y., Xu X., Jones J., Tiwari M., Ling J., Wang Y., Harismendy O., Sen G.L. HNRNPK maintains epidermal progenitor function through transcription of proliferation genes and degrading differentiation promoting mRNAs. Nat. Commun. 2019;10:4198. doi: 10.1038/s41467-019-12238-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shnyreva M., Schullery D.S., Suzuki H., Higaki Y., Bomsztyk K. Interaction of two multifunctional proteins. Heterogeneous nuclear ribonucleoprotein K and Y-box-binding protein. J. Biol. Chem. 2000;275:15498–15503. doi: 10.1074/jbc.275.20.15498. [DOI] [PubMed] [Google Scholar]
- 8.Samuel S.K., Spencer V.A., Bajno L., Sun J.M., Holth L.T., Oesterreich S., Davie J.R. In situ cross-linking by cisplatin of nuclear matrix-bound transcription factors to nuclear DNA of human breast cancer cells. Cancer Res. 1998;58:3004–3008. [PubMed] [Google Scholar]
- 9.Pintacuda G., Wei G., Roustan C., Kirmizitas B.A., Solcan N., Cerase A., Castello A., Mohammed S., Moindrot B., Nesterova T.B., Brockdorff N. hnRNPK recruits PCGF3/5-PRC1 to the xist RNA B-repeat to establish polycomb-mediated chromosomal silencing. Mol. Cell. 2017;68:955–969.e10. doi: 10.1016/j.molcel.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu Y., Wu W., Han Q., Wang Y., Li C., Zhang P., Xu H. New insights into the interplay between non-coding RNAs and RNA-binding protein HnRNPK in regulating cellular functions. Cells. 2019;8:62. doi: 10.3390/cells8010062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simko E.A.J., Liu H., Zhang T., Velasquez A., Teli S., Haeusler A.R., Wang J. G-quadruplexes offer a conserved structural motif for NONO recruitment to NEAT1 architectural lncRNA. Nucleic Acids Res. 2020;48:7421–7438. doi: 10.1093/nar/gkaa475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yano M., Okano H.J., Okano H. Involvement of Hu and heterogeneous nuclear ribonucleoprotein K in neuronal differentiation through p21 mRNA post-transcriptional regulation. J. Biol. Chem. 2005;280:12690–12699. doi: 10.1074/jbc.M411119200. [DOI] [PubMed] [Google Scholar]
- 13.Notari M., Neviani P., Santhanam R., Blaser B.W., Chang J.S., Galietta A., Willis A.E., Roy D.C., Caligiuri M.A., Marcucci G., Perrotti D. A MAPK/HNRPK pathway controls BCR/ABL oncogenic potential by regulating MYC mRNA translation. Blood. 2006;107:2507–2516. doi: 10.1182/blood-2005-09-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen T.M., Kabotyanski E.B., Reineke L.C., Shao J., Xiong F., Lee J.H., Dubrulle J., Johnson H., Stossi F., Tsoi P.S., et al. The SINEB1 element in the long non-coding RNA Malat1 is necessary for TDP-43 proteostasis. Nucleic Acids Res. 2020;48:2621–2642. doi: 10.1093/nar/gkz1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taiana E., Ronchetti D., Todoerti K., Nobili L., Tassone P., Amodio N., Neri A. LncRNA NEAT1 in paraspeckles: a structural scaffold for cellular DNA damage response systems? Noncoding RNA. 2020;6:26. doi: 10.3390/ncrna6030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamazaki T., Souquere S., Chujo T., Kobelke S., Chong Y.S., Fox A.H., Bond C.S., Nakagawa S., Pierron G., Hirose T. Functional domains of NEAT1 architectural lncRNA induce paraspeckle assembly through phase separation. Mol. Cell. 2018;70:1038–1053.e7. doi: 10.1016/j.molcel.2018.05.019. [DOI] [PubMed] [Google Scholar]
- 17.Naganuma T., Hirose T. Paraspeckle formation during the biogenesis of long non-coding RNAs. RNA Biol. 2013;10:456–461. doi: 10.4161/rna.23547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirose T., Nakagawa S. Paraspeckles: possible nuclear hubs by the RNA for the RNA. Biomol. Concepts. 2012;3:415–428. doi: 10.1515/bmc-2012-0017. [DOI] [PubMed] [Google Scholar]
- 19.Chung I.C., Chen L.C., Chung A.K., Chao M., Huang H.Y., Hsueh C., Tsang N.M., Chang K.P., Liang Y., Li H.P., Chang Y.S. Matrix metalloproteinase 12 is induced by heterogeneous nuclear ribonucleoprotein K and promotes migration and invasion in nasopharyngeal carcinoma. BMC Cancer. 2014;14:348. doi: 10.1186/1471-2407-14-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao R., Yu Y., Inoue A., Widodo N., Kaul S.C., Wadhwa R. Heterogeneous nuclear ribonucleoprotein K (hnRNP-K) promotes tumor metastasis by induction of genes involved in extracellular matrix, cell movement, and angiogenesis. J. Biol. Chem. 2013;288:15046–15056. doi: 10.1074/jbc.M113.466136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Y., Zeng Y., Xiao Z., Chen S., Li Y., Zou J., Zeng X. Role of heterogeneous nuclear ribonucleoprotein K in tumor development. J. Cell Biochem. 2019;120:14296–14305. doi: 10.1002/jcb.28867. [DOI] [PubMed] [Google Scholar]
- 22.Gallardo M., Hornbaker M.J., Zhang X., Hu P., Bueso-Ramos C., Post S.M. Aberrant hnRNP K expression: all roads lead to cancer. Cell Cycle. 2016;15:1552–1557. doi: 10.1080/15384101.2016.1164372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wen F., Shen A., Shanas R., Bhattacharyya A., Lian F., Hostetter G., Shi J. Higher expression of the heterogeneous nuclear ribonucleoprotein k in melanoma. Ann. Surg. Oncol. 2010;17:2619–2627. doi: 10.1245/s10434-010-1121-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong X., Song R., Song H., Zheng T., Wang J., Liang Y., Qi S., Lu Z., Song X., Jiang H., et al. PTEN antagonises Tcl1/hnRNPK-mediated G6PD pre-mRNA splicing which contributes to hepatocarcinogenesis. Gut. 2014;63:1635–1647. doi: 10.1136/gutjnl-2013-305302. [DOI] [PubMed] [Google Scholar]
- 25.Chen L.C., Chung I.C., Hsueh C., Tsang N.M., Chi L.M., Liang Y., Chen C.C., Wang L.J., Chang Y.S. The antiapoptotic protein, FLIP, is regulated by heterogeneous nuclear ribonucleoprotein K and correlates with poor overall survival of nasopharyngeal carcinoma patients. Cell Death Differ. 2010;17:1463–1473. doi: 10.1038/cdd.2010.24. [DOI] [PubMed] [Google Scholar]
- 26.Ciarlo M., Benelli R., Barbieri O., Minghelli S., Barboro P., Balbi C., Ferrari N. Regulation of neuroendocrine differentiation by AKT/hnRNPK/AR/beta-catenin signaling in prostate cancer cells. Int. J. Cancer. 2012;131:582–590. doi: 10.1002/ijc.26402. [DOI] [PubMed] [Google Scholar]
- 27.Wu C.S., Chang K.P., Chen L.C., Chen C.C., Liang Y., Hseuh C., Chang Y.S. Heterogeneous ribonucleoprotein K and thymidine phosphorylase are independent prognostic and therapeutic markers for oral squamous cell carcinoma. Oral Oncol. 2012;48:516–522. doi: 10.1016/j.oraloncology.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 28.Zhou R., Shanas R., Nelson M.A., Bhattacharyya A., Shi J. Increased expression of the heterogeneous nuclear ribonucleoprotein K in pancreatic cancer and its association with the mutant p53. Int. J. Cancer. 2010;126:395–404. doi: 10.1002/ijc.24744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Otoshi T., Tanaka T., Morimoto K., Nakatani T. Cytoplasmic accumulation of heterogeneous nuclear ribonucleoprotein K strongly promotes tumor invasion in renal cell carcinoma cells. PLoS One. 2015;10:e0145769. doi: 10.1371/journal.pone.0145769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Enge M., Bao W., Hedstrom E., Jackson S.P., Moumen A., Selivanova G. MDM2-dependent downregulation of p21 and hnRNP K provides a switch between apoptosis and growth arrest induced by pharmacologically activated p53. Cancer Cell. 2009;15:171–183. doi: 10.1016/j.ccr.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 31.Moumen A., Masterson P., O'Connor M.J., Jackson S.P. hnRNP K: an HDM2 target and transcriptional coactivator of p53 in response to DNA damage. Cell. 2005;123:1065–1078. doi: 10.1016/j.cell.2005.09.032. [DOI] [PubMed] [Google Scholar]
- 32.Lee S.W., Lee M.H., Park J.H., Kang S.H., Yoo H.M., Ka S.H., Oh Y.M., Jeon Y.J., Chung C.H. SUMOylation of hnRNP-K is required for p53-mediated cell-cycle arrest in response to DNA damage. EMBO J. 2012;31:4441–4452. doi: 10.1038/emboj.2012.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pelisch F., Pozzi B., Risso G., Munoz M.J., Srebrow A. DNA damage-induced heterogeneous nuclear ribonucleoprotein K sumoylation regulates p53 transcriptional activation. J. Biol. Chem. 2012;287:30789–30799. doi: 10.1074/jbc.M112.390120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gallardo M., Lee H.J., Zhang X., Bueso-Ramos C., Pageon L.R., McArthur M., Multani A., Nazha A., Manshouri T., Parker-Thornburg J., et al. hnRNP K is a haploinsufficient tumor suppressor that regulates proliferation and differentiation programs in hematologic malignancies. Cancer Cell. 2015;28:486–499. doi: 10.1016/j.ccell.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang Z., Ding G., Meng Z., Meng Q. The rational design of cell-penetrating peptides for application in delivery systems. Peptides. 2019;121:170149. doi: 10.1016/j.peptides.2019.170149. [DOI] [PubMed] [Google Scholar]
- 36.Kardani K., Milani A., S H.S., Bolhassani A. Cell penetrating peptides: the potent multi-cargo intracellular carriers. Expert Opin. Drug Deliv. 2019;16:1227–1258. doi: 10.1080/17425247.2019.1676720. [DOI] [PubMed] [Google Scholar]
- 37.Ruseska I., Zimmer A. Internalization mechanisms of cell-penetrating peptides. Beilstein J. Nanotechnol. 2020;11:101–123. doi: 10.3762/bjnano.11.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feng J., Xiao T., Lu S.S., Hung X.P., Yi H., He Q.Y., Huang W., Tang Y.Y., Xiao Z.Q. ANXA1derived peptides suppress gastric and colon cancer cell growth by targeting EphA2 degradation. Int. J. Oncol. 2020;57:1203–1213. doi: 10.3892/ijo.2020.5119. [DOI] [PubMed] [Google Scholar]
- 39.Thomas E., Dragojevic S., Price A., Raucher D. Thermally targeted p50 peptide inhibits proliferation and induces apoptosis of breast cancer cell lines. Macromol. Biosci. 2020;20:e2000170. doi: 10.1002/mabi.202000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jaraiz-Rodriguez M., Talaveron R., Garcia-Vicente L., Pelaz S.G., Dominguez-Prieto M., Alvarez-Vazquez A., Flores-Hernandez R., Sin W.C., Bechberger J., Medina J.M., et al. Connexin43 peptide, TAT-Cx43266-283, selectively targets glioma cells, impairs malignant growth, and enhances survival in mouse models in vivo. Neuro Oncol. 2020;22:493–504. doi: 10.1093/neuonc/noz243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alcalde J., Gonzalez-Munoz M., Villalobo A. Grb7-derived calmodulin-binding peptides inhibit proliferation, migration and invasiveness of tumor cells while they enhance attachment to the substrate. Heliyon. 2020;6:e03922. doi: 10.1016/j.heliyon.2020.e03922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Puvvula P.K., Moon A.M. Novel cell-penetrating peptides derived from scaffold-attachment- factor A inhibits cancer cell proliferation and survival. Front Oncol. 2021;11:621825. doi: 10.3389/fonc.2021.621825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Puvvula P.K., Yu Y., Sullivan K.R., Eyob H., Rosenberg J., Welm A., Huff C., Moon A.M. Inhibiting an RBM39/MLL1 epigenomic regulatory complex with dominant-negative peptides disrupts cancer cell transcription and proliferation. Cell Rep. 2021;35:109156. doi: 10.1016/j.celrep.2021.109156. [DOI] [PubMed] [Google Scholar]
- 44.Bomsztyk K., Denisenko O., Ostrowski J. hnRNP K: one protein multiple processes. Bioessays. 2004;26:629–638. doi: 10.1002/bies.20048. [DOI] [PubMed] [Google Scholar]
- 45.Barboro P., Ferrari N., Balbi C. Emerging roles of heterogeneous nuclear ribonucleoprotein K (hnRNP K) in cancer progression. Cancer Lett. 2014;352:152–159. doi: 10.1016/j.canlet.2014.06.019. [DOI] [PubMed] [Google Scholar]
- 46.Yin Z., Kobayashi M., Hu W., Higashi K., Begum N.A., Kurokawa K., Honjo T. RNA-binding motifs of hnRNP K are critical for induction of antibody diversification by activation-induced cytidine deaminase. Proc. Natl. Acad. Sci. U S A. 2020;117:11624–11635. doi: 10.1073/pnas.1921115117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fei T., Chen Y., Xiao T., Li W., Cato L., Zhang P., Cotter M.B., Bowden M., Lis R.T., Zhao S.G., et al. Genome-wide CRISPR screen identifies HNRNPL as a prostate cancer dependency regulating RNA splicing. Proc. Natl. Acad. Sci. U S A. 2017;114:E5207–E5215. doi: 10.1073/pnas.1617467114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen X., Gu P., Xie R., Han J., Liu H., Wang B., Xie W., Xie W., Zhong G., Chen C., et al. Heterogeneous nuclear ribonucleoprotein K is associated with poor prognosis and regulates proliferation and apoptosis in bladder cancer. J. Cell Mol. Med. 2017;21:1266–1279. doi: 10.1111/jcmm.12999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Derossi D., Chassaing G., Prochiantz A. Trojan peptides: the penetratin system for intracellular delivery. Trends Cell Biol. 1998;8:84–87. [PubMed] [Google Scholar]
- 50.Nakamoto M.Y., Lammer N.C., Batey R.T., Wuttke D.S. hnRNPK recognition of the B motif of Xist and other biological RNAs. Nucleic Acids Res. 2020;48:9320–9335. doi: 10.1093/nar/gkaa677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kiledjian M., Dreyfuss G. Primary structure and binding activity of the hnRNP U protein: binding RNA through RGG box. EMBO J. 1992;11:2655–2664. doi: 10.1002/j.1460-2075.1992.tb05331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ozdilek B.A., Thompson V.F., Ahmed N.S., White C.I., Batey R.T., Schwartz J.C. Intrinsically disordered RGG/RG domains mediate degenerate specificity in RNA binding. Nucleic Acids Res. 2017;45:7984–7996. doi: 10.1093/nar/gkx460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tang F., Li W., Chen Y., Wang D., Han J., Liu D. Downregulation of hnRNP K by RNAi inhibits growth of human lung carcinoma cells. Oncol. Lett. 2014;7:1073–1077. doi: 10.3892/ol.2014.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu Y., Li R., Zhang K., Wu W., Wang S., Zhang P., Xu H. The multifunctional RNA-binding protein hnRNPK is critical for the proliferation and differentiation of myoblasts. BMB Rep. 2018;51:350–355. doi: 10.5483/bmbrep.2018.51.7.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wells J., Graveel C.R., Bartley S.M., Madore S.J., Farnham P.J. The identification of E2F1-specific target genes. Proc. Natl. Acad. Sci. U S A. 2002;99:3890–3895. doi: 10.1073/pnas.062047499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Swiatkowska A., Dutkiewicz M., Machtel P., Janecki D.M., Kabacinska M., Zydowicz-Machtel P., Ciesiolka J. Regulation of the p53 expression profile by hnRNP K under stress conditions. RNA Biol. 2020;17:1402–1415. doi: 10.1080/15476286.2020.1771944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lubelsky Y., Ulitsky I. Sequences enriched in Alu repeats drive nuclear localization of long RNAs in human cells. Nature. 2018;555:107–111. doi: 10.1038/nature25757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shin C.H., Kim H.H. Functional roles of heterogeneous nuclear ribonucleoprotein K in post-transcriptional gene regulation. Precision Future Med. 2018;2:158–166. doi: 10.23838/pfm.2018.00107. [DOI] [Google Scholar]
- 59.Zarnack K., Konig J., Tajnik M., Martincorena I., Eustermann S., Stevant I., Reyes A., Anders S., Luscombe N.M., Ule J. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell. 2013;152:453–466. doi: 10.1016/j.cell.2012.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pfister A.S. Emerging role of the nucleolar stress response in autophagy. Front Cell Neurosci. 2019;13:156. doi: 10.3389/fncel.2019.00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lynch M., Chen L., Ravitz M.J., Mehtani S., Korenblat K., Pazin M.J., Schmidt E.V. hnRNP K binds a core polypyrimidine element in the eukaryotic translation initiation factor 4E (eIF4E) promoter, and its regulation of eIF4E contributes to neoplastic transformation. Mol. Cell Biol. 2005;25:6436–6453. doi: 10.1128/MCB.25.15.6436-6453.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Puvvula P.K., Desetty R.D., Pineau P., Marchio A., Moon A., Dejean A., Bischof O. Long noncoding RNA PANDA and scaffold-attachment-factor SAFA control senescence entry and exit. Nat. Commun. 2014;5:5323. doi: 10.1038/ncomms6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huarte M., Guttman M., Feldser D., Garber M., Koziol M.J., Kenzelmann-Broz D., Khalil A.M., Zuk O., Amit I., Rabani M., et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thompson P.J., Dulberg V., Moon K.M., Foster L.J., Chen C., Karimi M.M., Lorincz M.C. Correction: hnRNP K coordinates transcriptional silencing by SETDB1 in embryonic stem cells. PLoS Genet. 2016;12:e1006390. doi: 10.1371/journal.pgen.1006390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lu J., Gao F.H. Role and molecular mechanism of heterogeneous nuclear ribonucleoprotein K in tumor development and progression. Biomed. Rep. 2016;4:657–663. doi: 10.3892/br.2016.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Verchere J.F., Sauvage J.P., Rapaumbya G.R. Comparative study of various polyols as complexing agents for the acidimetric titration of tungstate. Analyst. 1990;115:637–640. doi: 10.1039/an9901500637. [DOI] [PubMed] [Google Scholar]
- 67.Wang B., Zhao M.Z., Cui N.P., Lin D.D., Zhang A.Y., Qin Y., Liu C.Y., Yan W.T., Shi J.H., Chen B.P. Kruppel-like factor 4 induces apoptosis and inhibits tumorigenic progression in SK-BR-3 breast cancer cells. FEBS Open Bio. 2015;5:147–154. doi: 10.1016/j.fob.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Preston G.A., Lyon T.T., Yin Y., Lang J.E., Solomon G., Annab L., Srinivasan D.G., Alcorta D.A., Barrett J.C. Induction of apoptosis by c-Fos protein. Mol. Cell Biol. 1996;16:211–218. doi: 10.1128/mcb.16.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stains J.P., Lecanda F., Towler D.A., Civitelli R. Heterogeneous nuclear ribonucleoprotein K represses transcription from a cytosine/thymidine-rich element in the osteocalcin promoter. Biochem. J. 2005;385:613–623. doi: 10.1042/BJ20040680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim J., Venkata N.C., Hernandez Gonzalez G.A., Khanna N., Belmont A.S. Gene expression amplification by nuclear speckle association. J. Cell Biol. 2020;219:e201904046. doi: 10.1083/jcb.201904046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.West J.A., Davis C.P., Sunwoo H., Simon M.D., Sadreyev R.I., Wang P.I., Tolstorukov M.Y., Kingston R.E. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol. Cell. 2014;55:791–802. doi: 10.1016/j.molcel.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bao X., Wu H., Zhu X., Guo X., Hutchins A.P., Luo Z., Song H., Chen Y., Lai K., Yin M., et al. The p53-induced lincRNA-p21 derails somatic cell reprogramming by sustaining H3K9me3 and CpG methylation at pluripotency gene promoters. Cell Res. 2015;25:80–92. doi: 10.1038/cr.2014.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained in the article and supplemental data files.