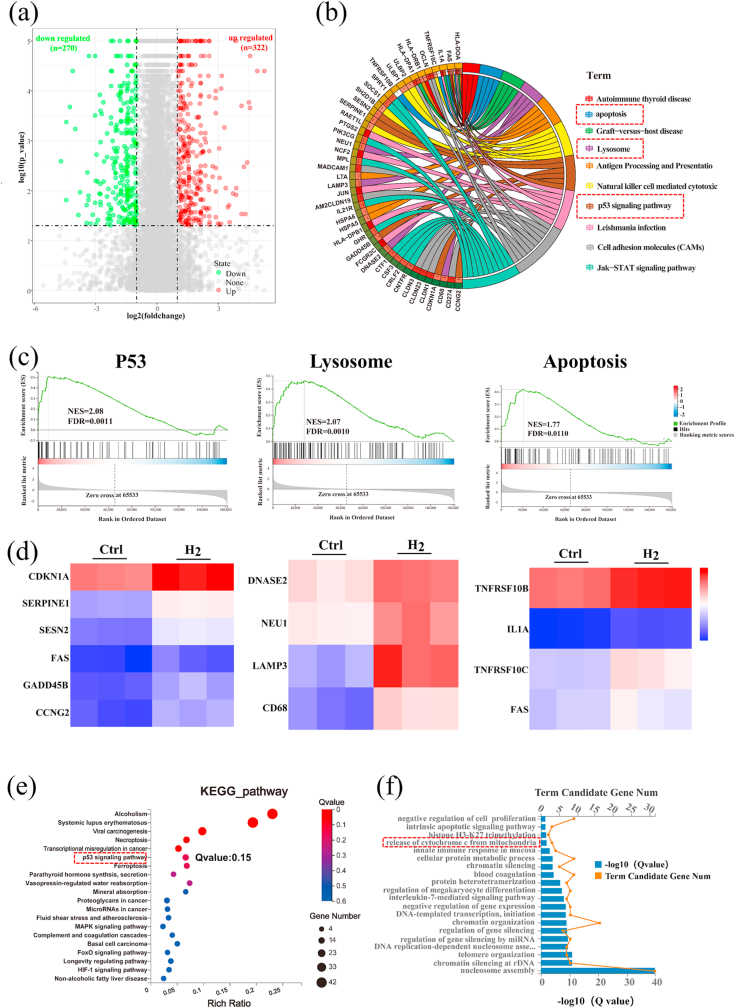
Fig. 3.
Hydrogen regulated gene expressions related to the P53-mediated apoptosis signaling pathway. (a) A volcano plot showing up/down-regulated genes after M − H2 treatment vs. the Ctrl group. Genes with an absolute fold change of >2 and a P value of <0.05 are highlighted in green and red color separately, denoting the down- and up-regulated genes, respectively. (b) Circular visualization of the gene-annotation enrichment analysis after gene set enrichment analysis (GSEA) was conducted using the Molecular Signatures Database (MSigDB). (c) The gene set enrichment analysis of the P53, lysosome and apoptosis pathway. (NES: normalized enrichment score). (d) A heat map of differentially expressed genes associated with three apoptotic pathways after H2 treatment, a fold change of >2 and a P value of <0.05 compared to the Ctrl group was analyzed. (e, f) An analysis of the enriched differentially expressed genes using the Kyoto Encyclopedia of Genes (KEGG) and Genomes (GO) database.