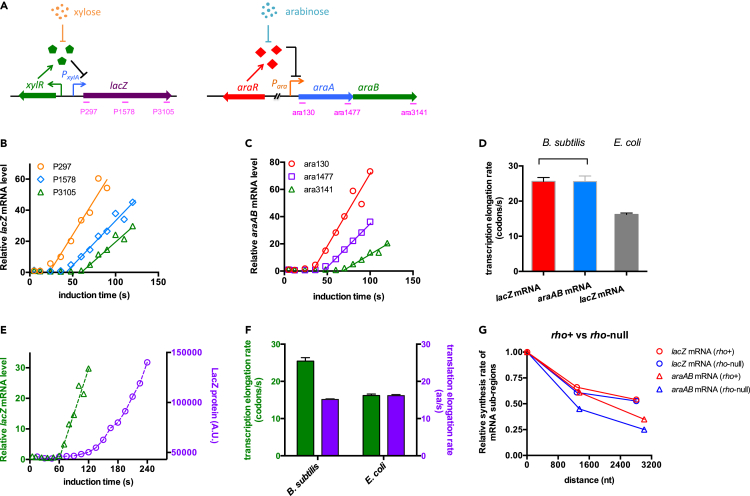
Figure 1.
Characterization of transcription and translation kinetics of Bacillus subtilis
Cells were grown in glycerol plus casamino acid (gly + cAA) medium.
(A) Two inducible systems used in this study. A xylR-PxylA-lacZ cassette was integrated into the lacA locus of B. subtilis genome. The expression of lacZ gene is controlled by the XylR repressor and thus is induced by xylose. The other inducible system is the native ara operon of B. subtilis, which is controlled by AraR and is induced by arabinose. We focused on the araAB region of the ara operon. Regions detected by three qRT-PCR primers are labeled in pink.
(B) The induction kinetics of the lacZ mRNA. Three pairs of primers were used to detect the abundances of lacZ mRNA sub-regions. The rising part of each induction curve was fitted to a linear line.
(C) The induction kinetics of araAB mRNA detected by three pairs of qRT-PCR primers, being similar to (B)
(D) The transcription elongation rates of both B. subtilis and E. coli. Data of E. coli lacZ mRNA are from Zhu et al (2019). Data are represented as mean ± SD.
(E) The induction curve of the intact lacZ mRNA (detected by P3105, green triangles) is plotted together with the induction curve of LacZ protein (purple circles).
(F) The transcription and translation elongation rates of B. subtilis and E. coli. Data of E. coli cells are from Zhu et al (2019). Data are represented as mean ± SD.
(G) The transcription processivity of lacZ mRNA and araAB mRNA in both rho+ (red symbols) and rho-null strains (blue symbols) of B. subtilis. The relative accumulation rates of mRNA sub-regions (slope of the linear induction curve in B and C) are plotted against the hybridization locations of primers in lacZ or araAB mRNA. The detection positions of the 5' primer (P297 for lacZ mRNA and ara130 for araAB mRNA) were set as location “zero” (x axis). The accumulation rates of the 5′ mRNA sub-regions detected by P297 or ara130 were set as “1” (y axis).