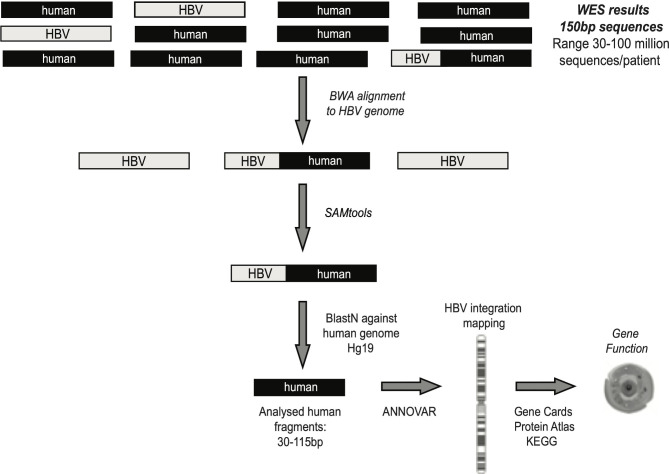
Figure 1.
Bioinformatic workflow for the identification of chimeric HBV human sequences. Following quality control (by Trimmomatic), sequencing reads were aligned to virus-specific genome by Burrows Wheeler Aligner-Maximal Exaxt Matches (BWA-MEM) in order to extract all reads that contained HBV fragments. SAMtools software was applied to extrapolate all chimeric HBV human sequences, representing the HBV integrations into human genome. ANNOVAR software was used to map HBV integrations at chromosome and gene level. The functionality of genes involved in HBV integration was retrieved by Gene Cards, Protein Atlas and KEGG databases. WES, whole exome sequencing.