Extended Data Fig. 7 |. GSDM acidic patches and their mutations.
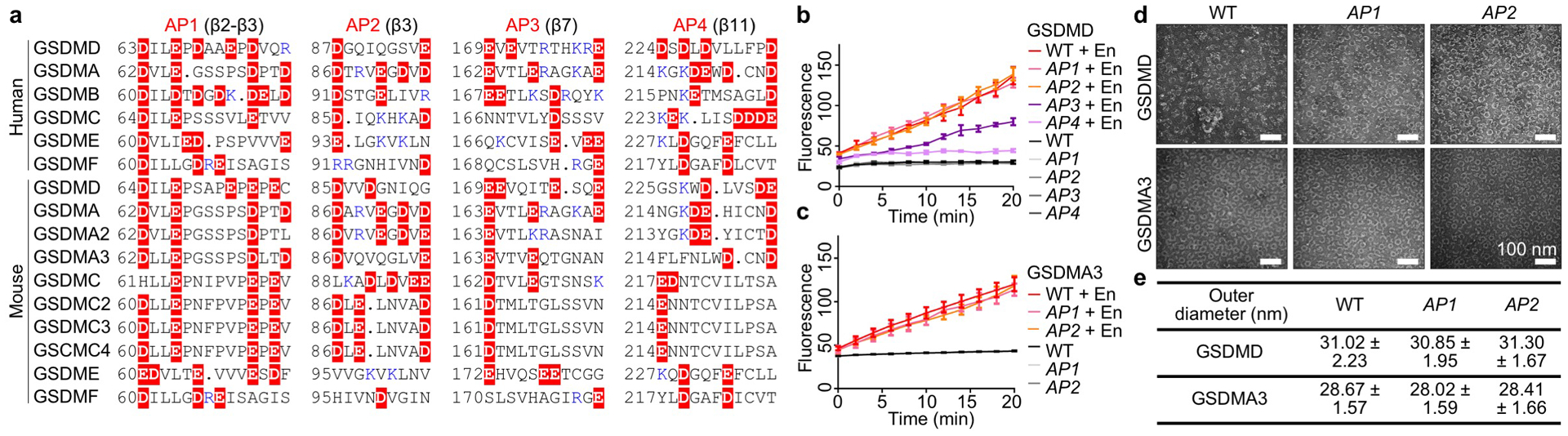
a, Locations of APs shown by aligned GSDM sequences. Dots: Strings of omitted uncharged residues. Red highlight: Acidic residues. Blue: Basic residues. Of note, the basic residues near the APs may face the membrane (such as those in BP3) and therefore do not necessarily weaken the acidity of the pore conduit. b, Assessment of alanine mutations of GSDMD APs 1 through 4 by Tb3+ leakage (n = 3 biological replicates). c, Assessment of alanine mutations of GSDMA3 AP1 and AP2 by Tb3+ liposome leakage (n = 3 biological replicates). d, Negative-staining EM images of WT and AP-mutant GSDMD and GSDMA3 assemblies, solubilized from liposomes using C12E8 and cholate, respectively. All scale bars: 100 nm. e, Outer diameters of WT or AP-mutant GSDMD and GSDMA3 assemblies, measured under negative-staining EM (n = 50 particles per group). Data shown in b, c, and e are mean ± s.d.. Data shown in d are representative of three independent experiments.
