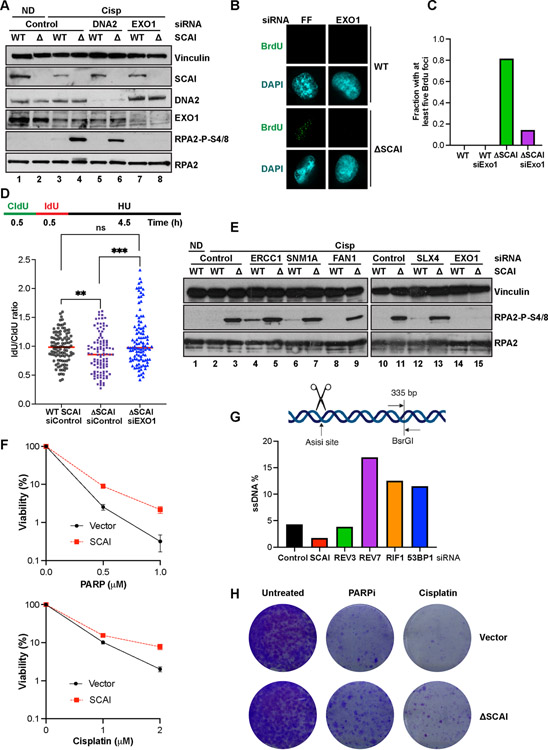
Figure 5. SCAI protects stalled forks but not DSBs against resection by EXO1 and promotes resistance to PARP inhibitors.
(A) Cells treated with the indicated siRNA for 72 h were treated with vehicle or 2 mM cisp for 16 h and analyzed by immunoblotting. (B) IF analyses showing representative BrdU foci from WT and SCAI nulls following 60 h treatment with the indicated siRNAs. Cells were treated with MMC for 6 h prior to IF. (C) Quantification of B. (D) Top. Schematic showing DNA fiber assay protocol. Cells were pulsed with CldU followed by IdU for 30 m each after which forks were stalled by 4 mM HU treatment for 4.5 h. Bottom. WT and SCAI-nulls were treated with indicated siRNAs for 60 h before labeling as in schematic. DNA combing analyses was performed and approximately 100 fibers were quantified and plotted. Mann Whitney test, ns=p>0.05, **=p<0.01, ***=p<0.001. (E) WT and SCAI-null U2OS were treated with the indicated siRNAs for 72 h, then treated with vehicle (ND) or 2 μM cisp for 18 h before immunoblotting. The same samples were run in lanes 2–3 and lanes 10–11. Depletion of indicated proteins is shown in Figure S5F. (F) ER-AsiSi U2OS transfected with the indicated siRNAs for 60 h were treated with Tamoxifen to induce DSBs. Cells were harvested in low-melting agarose, proteinase-treated and genomic DNA was extracted. After restriction digest, qPCR was performed to determine resection efficiency. (G) BRCA1-null, TP53-null RPE1 cells expressing SCAI or vector control were treated with vehicle, or the indicated doses of olaparib or cisp for 2 weeks and allowed to form colonies. Mean ± SD two independent experiments. Representative images shown.