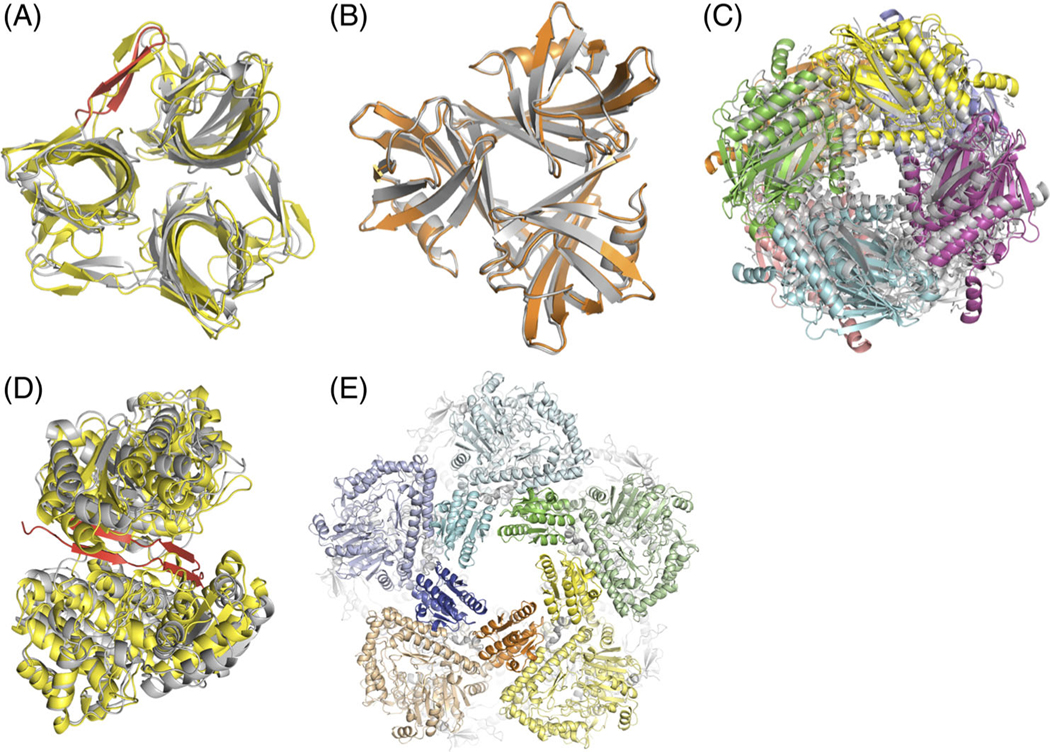
FIGURE 1.
A, T110: Our best model (** rating) of the fiber head domain of raptor adenovirus 1 (yellow) superimposed on the crystal structure (gray). Predicting and modeling the beta-hairpin in the native structure (red) was crucial to prediction success. The Lrmsd of 2.35 Å was the best among all groups. B, T111: Our high-quality model (***) of the fiber head domain of lizard adenovirus 2 (orange) superimposed on the crystal structure of a close homolog (gray), snake adenovirus 1 fiber head. C, T118: Our best model (**) of fructose 1,6-bisphosphatase (color) superimposed on the crystal structure of a close homolog (gray). D, T119: Our best model (*) of alcohol dehydrogenase dimer (yellow) superimposed on the crystal structure (gray). The model is missing cross-beta interactions between the subunits at the interface (red). E, T136: Our model (**) of LdcA decamer where the five subunits on top are displayed in different colors. For each subunit, the wing domains are highlighted in brighter hues. The Lrmsd of 2.44 Å was the best among all groups