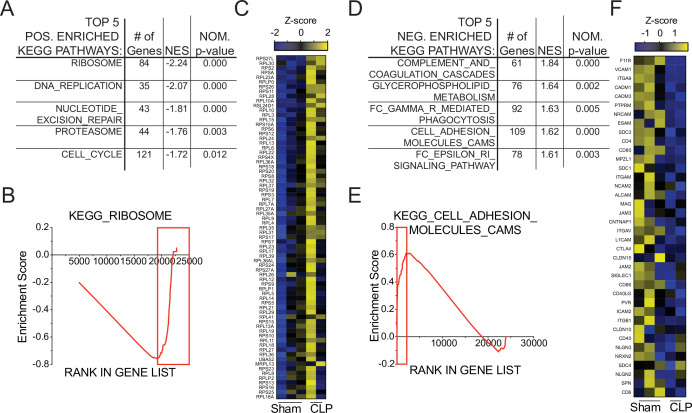
Figure 6. Gene set enrichment analysis (GSEA) reveals long-term sepsis-induced differences in molecular pathways of pre-existing memory CD8 T cells.
Top 5 KEGG pathways positively- (A) and negatively- (D) enriched in CLP hosts. Enrichment scores for Ribosomal- (B) and Adhesion- (E) associated genes. Red box indicates leading edge of enriched region; genes enriched in CLP - box to right, genes enriched in Sham – box to left. Gene expression heatmap of core enriched genes for Ribosomal (C) and Adhesion (F) associated genes.